
Enterococcus pallens ATCC BAA-351
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Enterococcaceae; Enterococcus; Enterococcus pallens
Average proteome isoelectric point is 6.05
Get precalculated fractions of proteins
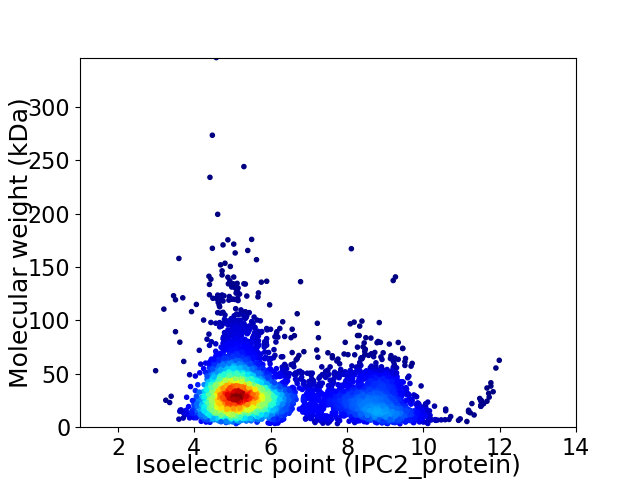
Virtual 2D-PAGE plot for 5181 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R2SC12|R2SC12_9ENTE DNA translocase FtsK OS=Enterococcus pallens ATCC BAA-351 OX=1158607 GN=UAU_02702 PE=4 SV=1
MM1 pKa = 7.09YY2 pKa = 10.42KK3 pKa = 10.13SGKK6 pKa = 8.46HH7 pKa = 4.61WMFATITAFTGVLGASTLVTTEE29 pKa = 3.9AQATSASQDD38 pKa = 3.57DD39 pKa = 4.06AGMEE43 pKa = 4.13IDD45 pKa = 3.64TGTDD49 pKa = 2.78QVLANQEE56 pKa = 4.28TASIPAPEE64 pKa = 4.18TQEE67 pKa = 4.23AVNPDD72 pKa = 3.74PVVDD76 pKa = 3.89NTPEE80 pKa = 3.87SEE82 pKa = 4.34APVSEE87 pKa = 4.2IASEE91 pKa = 4.48SVVQTEE97 pKa = 4.66LEE99 pKa = 4.29STTEE103 pKa = 4.08STSTTEE109 pKa = 4.21SEE111 pKa = 4.64SNMDD115 pKa = 3.44SASISEE121 pKa = 4.34SVSVSEE127 pKa = 4.67SMSISEE133 pKa = 4.95SEE135 pKa = 4.21SSSQSRR141 pKa = 11.84SEE143 pKa = 4.04EE144 pKa = 3.97TSLSISTSEE153 pKa = 4.11STSNSALAEE162 pKa = 4.37VNNSASNSLSEE173 pKa = 4.41STSHH177 pKa = 6.71SEE179 pKa = 4.66SASTWVSNSLSEE191 pKa = 4.25STSNSEE197 pKa = 3.42NRR199 pKa = 11.84SAVNSLNKK207 pKa = 9.97EE208 pKa = 3.84KK209 pKa = 10.84SKK211 pKa = 11.05QPTEE215 pKa = 4.28KK216 pKa = 10.18EE217 pKa = 3.68QSANEE222 pKa = 3.96TNKK225 pKa = 10.74AKK227 pKa = 10.71DD228 pKa = 3.71LFDD231 pKa = 4.29QIVPDD236 pKa = 3.83GTEE239 pKa = 3.74VKK241 pKa = 10.49VDD243 pKa = 3.21ARR245 pKa = 11.84SNEE248 pKa = 4.06LVFTMKK254 pKa = 10.72DD255 pKa = 3.02GTVPSNALIHH265 pKa = 6.69AAQKK269 pKa = 9.82YY270 pKa = 9.12AAANDD275 pKa = 4.64LIAYY279 pKa = 7.8FSVRR283 pKa = 11.84GANGRR288 pKa = 11.84SAKK291 pKa = 10.25INVSADD297 pKa = 3.26PDD299 pKa = 3.76VNRR302 pKa = 11.84EE303 pKa = 4.04DD304 pKa = 5.23FEE306 pKa = 5.19DD307 pKa = 4.95AINNPGTSNSPTSVNVAVPDD327 pKa = 3.64QVLNDD332 pKa = 3.74ATHH335 pKa = 6.79KK336 pKa = 11.06ANAIANGTYY345 pKa = 10.83AKK347 pKa = 9.36VTTYY351 pKa = 11.37DD352 pKa = 3.44EE353 pKa = 5.64LRR355 pKa = 11.84AAWSNASITYY365 pKa = 9.9IDD367 pKa = 3.61VTAKK371 pKa = 8.72ITYY374 pKa = 10.06SSGAMSARR382 pKa = 11.84ANGASVIVQGNNNIIDD398 pKa = 4.67LGTQTFNWGGITSSTTFTLSNVIAQQGFGGNDD430 pKa = 2.91GNAYY434 pKa = 10.73SLINPSSATQLTANVNNVTLTKK456 pKa = 10.38SDD458 pKa = 3.86SNGNNPIHH466 pKa = 6.85LMYY469 pKa = 10.82GVGSKK474 pKa = 10.62LVFSGTNVFNISNEE488 pKa = 3.54ITRR491 pKa = 11.84SVGSINFANNSSVTLNRR508 pKa = 11.84TSNDD512 pKa = 2.66IAFSQFYY519 pKa = 10.73FEE521 pKa = 4.23TRR523 pKa = 11.84AAAGSVGYY531 pKa = 10.57GNSITMGDD539 pKa = 3.68GSSNTARR546 pKa = 11.84TYY548 pKa = 10.93NGQSANYY555 pKa = 7.72PAVYY559 pKa = 10.92YY560 pKa = 9.67MIDD563 pKa = 3.98KK564 pKa = 9.22ITVGDD569 pKa = 3.79NCNWTQTGFQYY580 pKa = 10.55FINGQQLASSSATYY594 pKa = 10.21TFGQNFNLSIPQSTQPGAIRR614 pKa = 11.84IQGTQKK620 pKa = 10.27VVFNAGTTLDD630 pKa = 3.51INQWASGAVIQMNSSGSSVTFISPKK655 pKa = 8.2QLHH658 pKa = 5.55MQVSTSSGAPNTGPLISGAGTFTMNNSQISTWQGTDD694 pKa = 3.02SQAATPGGNRR704 pKa = 11.84NLKK707 pKa = 9.6FVTMTIRR714 pKa = 11.84NGTTTITDD722 pKa = 3.24VGGNTTSSNIIDD734 pKa = 3.52SSTRR738 pKa = 11.84EE739 pKa = 3.85LATTAIEE746 pKa = 4.39PGTVKK751 pKa = 10.43VNYY754 pKa = 9.15VDD756 pKa = 3.69QYY758 pKa = 10.21GNVVKK763 pKa = 10.01TIDD766 pKa = 3.46VPIDD770 pKa = 3.26KK771 pKa = 10.24DD772 pKa = 3.69VNYY775 pKa = 9.77IGQYY779 pKa = 9.64IDD781 pKa = 5.54LVNKK785 pKa = 10.37DD786 pKa = 3.68YY787 pKa = 11.68ALTNMPANYY796 pKa = 8.24MWAIASQVPPSALTDD811 pKa = 3.65KK812 pKa = 11.17QSGGDD817 pKa = 3.55NTTSADD823 pKa = 3.62DD824 pKa = 3.74GDD826 pKa = 5.15AYY828 pKa = 10.74GQANVAIVPMEE839 pKa = 4.0GTEE842 pKa = 3.93YY843 pKa = 10.16TYY845 pKa = 11.08NIYY848 pKa = 10.82VYY850 pKa = 10.13GQANNSVQYY859 pKa = 10.52QYY861 pKa = 11.88VDD863 pKa = 2.91AKK865 pKa = 9.53TGLVVSSDD873 pKa = 3.8YY874 pKa = 11.27LGSGQEE880 pKa = 3.88ASGTNKK886 pKa = 9.22TPANYY891 pKa = 10.64GNVIDD896 pKa = 4.49WTNSYY901 pKa = 8.14YY902 pKa = 10.96TNDD905 pKa = 3.51LVADD909 pKa = 5.49GYY911 pKa = 11.25HH912 pKa = 6.01YY913 pKa = 10.73ATGSEE918 pKa = 4.41LNGFTQATTTTVGTDD933 pKa = 3.31PTLVTIYY940 pKa = 10.48VAADD944 pKa = 3.42VQTVNVNFVNSDD956 pKa = 3.32GTPLVGQAANVSINGTSGAEE976 pKa = 3.61MSYY979 pKa = 11.72ADD981 pKa = 5.58LISGYY986 pKa = 8.22LTQAGYY992 pKa = 9.47HH993 pKa = 6.33ADD995 pKa = 3.5VNGVFTFDD1003 pKa = 3.38FTANGTSNTDD1013 pKa = 3.34SNPQSLTVTLYY1024 pKa = 10.3PDD1026 pKa = 3.33YY1027 pKa = 11.03QLAGVVGNNQPASDD1041 pKa = 4.22PATGAAYY1048 pKa = 8.33EE1049 pKa = 3.99QLPTSVTEE1057 pKa = 4.03GNYY1060 pKa = 10.11GVAGVSNGAINLGVTDD1076 pKa = 5.15ADD1078 pKa = 4.14LARR1081 pKa = 11.84NGYY1084 pKa = 9.41IYY1086 pKa = 10.44TVTGPDD1092 pKa = 3.3GQTYY1096 pKa = 7.91ATLAEE1101 pKa = 4.35ALAANPNFDD1110 pKa = 3.5TTTNGAGSVDD1120 pKa = 3.58NTPQGFVVNYY1130 pKa = 10.25DD1131 pKa = 3.25VDD1133 pKa = 4.06PVSEE1137 pKa = 4.3STSMSEE1143 pKa = 4.36SEE1145 pKa = 4.43SLSSSVSDD1153 pKa = 3.98SEE1155 pKa = 4.44SDD1157 pKa = 3.58STSTSISDD1165 pKa = 3.86SEE1167 pKa = 4.4SDD1169 pKa = 3.61STSTSISDD1177 pKa = 3.65SEE1179 pKa = 4.42STSRR1183 pKa = 11.84SEE1185 pKa = 5.6DD1186 pKa = 3.36STSDD1190 pKa = 3.85SISSSISEE1198 pKa = 4.38SEE1200 pKa = 4.5SISTSDD1206 pKa = 4.64SISDD1210 pKa = 4.09SISDD1214 pKa = 4.05SEE1216 pKa = 4.77SDD1218 pKa = 3.61SLSNSISDD1226 pKa = 4.29SISDD1230 pKa = 4.05SEE1232 pKa = 4.77SDD1234 pKa = 3.61SLSNSISDD1242 pKa = 4.54SISDD1246 pKa = 3.79STSDD1250 pKa = 4.35SISDD1254 pKa = 4.29SISDD1258 pKa = 4.46SISDD1262 pKa = 3.79STSDD1266 pKa = 4.35SISDD1270 pKa = 4.29SISDD1274 pKa = 4.41SISDD1278 pKa = 3.77SLSNSISDD1286 pKa = 4.54SISDD1290 pKa = 3.79STSDD1294 pKa = 4.35SISDD1298 pKa = 4.29SISDD1302 pKa = 4.28SISSSEE1308 pKa = 4.19SLSEE1312 pKa = 4.63SISEE1316 pKa = 4.19SQSLSISISEE1326 pKa = 4.26SDD1328 pKa = 3.63STSDD1332 pKa = 4.25SISDD1336 pKa = 3.98SEE1338 pKa = 4.52SDD1340 pKa = 4.25SISDD1344 pKa = 4.43SISDD1348 pKa = 4.46SISDD1352 pKa = 3.79STSDD1356 pKa = 4.02SISDD1360 pKa = 3.85SEE1362 pKa = 4.45SLSEE1366 pKa = 4.23SVSEE1370 pKa = 4.46SEE1372 pKa = 4.56SLSISISEE1380 pKa = 4.26SDD1382 pKa = 3.6STSDD1386 pKa = 3.51SLSDD1390 pKa = 3.91SEE1392 pKa = 5.25SDD1394 pKa = 3.94SISNSISDD1402 pKa = 4.81SISDD1406 pKa = 3.67STSNSISDD1414 pKa = 3.92SEE1416 pKa = 4.43SLSEE1420 pKa = 4.23SVSEE1424 pKa = 4.46SEE1426 pKa = 4.56SLSISISEE1434 pKa = 4.26SDD1436 pKa = 3.6STSDD1440 pKa = 3.51SLSDD1444 pKa = 3.91SEE1446 pKa = 4.94SDD1448 pKa = 4.24SISDD1452 pKa = 4.43SISDD1456 pKa = 4.51SISSSISNSDD1466 pKa = 3.5SLSEE1470 pKa = 4.16SVSKK1474 pKa = 11.08SEE1476 pKa = 4.38SLSISISEE1484 pKa = 4.33STSTSTSISDD1494 pKa = 3.58SSSTSTSTSNSDD1506 pKa = 3.24SDD1508 pKa = 4.02SLSNSISDD1516 pKa = 3.81SRR1518 pKa = 11.84SEE1520 pKa = 3.83
MM1 pKa = 7.09YY2 pKa = 10.42KK3 pKa = 10.13SGKK6 pKa = 8.46HH7 pKa = 4.61WMFATITAFTGVLGASTLVTTEE29 pKa = 3.9AQATSASQDD38 pKa = 3.57DD39 pKa = 4.06AGMEE43 pKa = 4.13IDD45 pKa = 3.64TGTDD49 pKa = 2.78QVLANQEE56 pKa = 4.28TASIPAPEE64 pKa = 4.18TQEE67 pKa = 4.23AVNPDD72 pKa = 3.74PVVDD76 pKa = 3.89NTPEE80 pKa = 3.87SEE82 pKa = 4.34APVSEE87 pKa = 4.2IASEE91 pKa = 4.48SVVQTEE97 pKa = 4.66LEE99 pKa = 4.29STTEE103 pKa = 4.08STSTTEE109 pKa = 4.21SEE111 pKa = 4.64SNMDD115 pKa = 3.44SASISEE121 pKa = 4.34SVSVSEE127 pKa = 4.67SMSISEE133 pKa = 4.95SEE135 pKa = 4.21SSSQSRR141 pKa = 11.84SEE143 pKa = 4.04EE144 pKa = 3.97TSLSISTSEE153 pKa = 4.11STSNSALAEE162 pKa = 4.37VNNSASNSLSEE173 pKa = 4.41STSHH177 pKa = 6.71SEE179 pKa = 4.66SASTWVSNSLSEE191 pKa = 4.25STSNSEE197 pKa = 3.42NRR199 pKa = 11.84SAVNSLNKK207 pKa = 9.97EE208 pKa = 3.84KK209 pKa = 10.84SKK211 pKa = 11.05QPTEE215 pKa = 4.28KK216 pKa = 10.18EE217 pKa = 3.68QSANEE222 pKa = 3.96TNKK225 pKa = 10.74AKK227 pKa = 10.71DD228 pKa = 3.71LFDD231 pKa = 4.29QIVPDD236 pKa = 3.83GTEE239 pKa = 3.74VKK241 pKa = 10.49VDD243 pKa = 3.21ARR245 pKa = 11.84SNEE248 pKa = 4.06LVFTMKK254 pKa = 10.72DD255 pKa = 3.02GTVPSNALIHH265 pKa = 6.69AAQKK269 pKa = 9.82YY270 pKa = 9.12AAANDD275 pKa = 4.64LIAYY279 pKa = 7.8FSVRR283 pKa = 11.84GANGRR288 pKa = 11.84SAKK291 pKa = 10.25INVSADD297 pKa = 3.26PDD299 pKa = 3.76VNRR302 pKa = 11.84EE303 pKa = 4.04DD304 pKa = 5.23FEE306 pKa = 5.19DD307 pKa = 4.95AINNPGTSNSPTSVNVAVPDD327 pKa = 3.64QVLNDD332 pKa = 3.74ATHH335 pKa = 6.79KK336 pKa = 11.06ANAIANGTYY345 pKa = 10.83AKK347 pKa = 9.36VTTYY351 pKa = 11.37DD352 pKa = 3.44EE353 pKa = 5.64LRR355 pKa = 11.84AAWSNASITYY365 pKa = 9.9IDD367 pKa = 3.61VTAKK371 pKa = 8.72ITYY374 pKa = 10.06SSGAMSARR382 pKa = 11.84ANGASVIVQGNNNIIDD398 pKa = 4.67LGTQTFNWGGITSSTTFTLSNVIAQQGFGGNDD430 pKa = 2.91GNAYY434 pKa = 10.73SLINPSSATQLTANVNNVTLTKK456 pKa = 10.38SDD458 pKa = 3.86SNGNNPIHH466 pKa = 6.85LMYY469 pKa = 10.82GVGSKK474 pKa = 10.62LVFSGTNVFNISNEE488 pKa = 3.54ITRR491 pKa = 11.84SVGSINFANNSSVTLNRR508 pKa = 11.84TSNDD512 pKa = 2.66IAFSQFYY519 pKa = 10.73FEE521 pKa = 4.23TRR523 pKa = 11.84AAAGSVGYY531 pKa = 10.57GNSITMGDD539 pKa = 3.68GSSNTARR546 pKa = 11.84TYY548 pKa = 10.93NGQSANYY555 pKa = 7.72PAVYY559 pKa = 10.92YY560 pKa = 9.67MIDD563 pKa = 3.98KK564 pKa = 9.22ITVGDD569 pKa = 3.79NCNWTQTGFQYY580 pKa = 10.55FINGQQLASSSATYY594 pKa = 10.21TFGQNFNLSIPQSTQPGAIRR614 pKa = 11.84IQGTQKK620 pKa = 10.27VVFNAGTTLDD630 pKa = 3.51INQWASGAVIQMNSSGSSVTFISPKK655 pKa = 8.2QLHH658 pKa = 5.55MQVSTSSGAPNTGPLISGAGTFTMNNSQISTWQGTDD694 pKa = 3.02SQAATPGGNRR704 pKa = 11.84NLKK707 pKa = 9.6FVTMTIRR714 pKa = 11.84NGTTTITDD722 pKa = 3.24VGGNTTSSNIIDD734 pKa = 3.52SSTRR738 pKa = 11.84EE739 pKa = 3.85LATTAIEE746 pKa = 4.39PGTVKK751 pKa = 10.43VNYY754 pKa = 9.15VDD756 pKa = 3.69QYY758 pKa = 10.21GNVVKK763 pKa = 10.01TIDD766 pKa = 3.46VPIDD770 pKa = 3.26KK771 pKa = 10.24DD772 pKa = 3.69VNYY775 pKa = 9.77IGQYY779 pKa = 9.64IDD781 pKa = 5.54LVNKK785 pKa = 10.37DD786 pKa = 3.68YY787 pKa = 11.68ALTNMPANYY796 pKa = 8.24MWAIASQVPPSALTDD811 pKa = 3.65KK812 pKa = 11.17QSGGDD817 pKa = 3.55NTTSADD823 pKa = 3.62DD824 pKa = 3.74GDD826 pKa = 5.15AYY828 pKa = 10.74GQANVAIVPMEE839 pKa = 4.0GTEE842 pKa = 3.93YY843 pKa = 10.16TYY845 pKa = 11.08NIYY848 pKa = 10.82VYY850 pKa = 10.13GQANNSVQYY859 pKa = 10.52QYY861 pKa = 11.88VDD863 pKa = 2.91AKK865 pKa = 9.53TGLVVSSDD873 pKa = 3.8YY874 pKa = 11.27LGSGQEE880 pKa = 3.88ASGTNKK886 pKa = 9.22TPANYY891 pKa = 10.64GNVIDD896 pKa = 4.49WTNSYY901 pKa = 8.14YY902 pKa = 10.96TNDD905 pKa = 3.51LVADD909 pKa = 5.49GYY911 pKa = 11.25HH912 pKa = 6.01YY913 pKa = 10.73ATGSEE918 pKa = 4.41LNGFTQATTTTVGTDD933 pKa = 3.31PTLVTIYY940 pKa = 10.48VAADD944 pKa = 3.42VQTVNVNFVNSDD956 pKa = 3.32GTPLVGQAANVSINGTSGAEE976 pKa = 3.61MSYY979 pKa = 11.72ADD981 pKa = 5.58LISGYY986 pKa = 8.22LTQAGYY992 pKa = 9.47HH993 pKa = 6.33ADD995 pKa = 3.5VNGVFTFDD1003 pKa = 3.38FTANGTSNTDD1013 pKa = 3.34SNPQSLTVTLYY1024 pKa = 10.3PDD1026 pKa = 3.33YY1027 pKa = 11.03QLAGVVGNNQPASDD1041 pKa = 4.22PATGAAYY1048 pKa = 8.33EE1049 pKa = 3.99QLPTSVTEE1057 pKa = 4.03GNYY1060 pKa = 10.11GVAGVSNGAINLGVTDD1076 pKa = 5.15ADD1078 pKa = 4.14LARR1081 pKa = 11.84NGYY1084 pKa = 9.41IYY1086 pKa = 10.44TVTGPDD1092 pKa = 3.3GQTYY1096 pKa = 7.91ATLAEE1101 pKa = 4.35ALAANPNFDD1110 pKa = 3.5TTTNGAGSVDD1120 pKa = 3.58NTPQGFVVNYY1130 pKa = 10.25DD1131 pKa = 3.25VDD1133 pKa = 4.06PVSEE1137 pKa = 4.3STSMSEE1143 pKa = 4.36SEE1145 pKa = 4.43SLSSSVSDD1153 pKa = 3.98SEE1155 pKa = 4.44SDD1157 pKa = 3.58STSTSISDD1165 pKa = 3.86SEE1167 pKa = 4.4SDD1169 pKa = 3.61STSTSISDD1177 pKa = 3.65SEE1179 pKa = 4.42STSRR1183 pKa = 11.84SEE1185 pKa = 5.6DD1186 pKa = 3.36STSDD1190 pKa = 3.85SISSSISEE1198 pKa = 4.38SEE1200 pKa = 4.5SISTSDD1206 pKa = 4.64SISDD1210 pKa = 4.09SISDD1214 pKa = 4.05SEE1216 pKa = 4.77SDD1218 pKa = 3.61SLSNSISDD1226 pKa = 4.29SISDD1230 pKa = 4.05SEE1232 pKa = 4.77SDD1234 pKa = 3.61SLSNSISDD1242 pKa = 4.54SISDD1246 pKa = 3.79STSDD1250 pKa = 4.35SISDD1254 pKa = 4.29SISDD1258 pKa = 4.46SISDD1262 pKa = 3.79STSDD1266 pKa = 4.35SISDD1270 pKa = 4.29SISDD1274 pKa = 4.41SISDD1278 pKa = 3.77SLSNSISDD1286 pKa = 4.54SISDD1290 pKa = 3.79STSDD1294 pKa = 4.35SISDD1298 pKa = 4.29SISDD1302 pKa = 4.28SISSSEE1308 pKa = 4.19SLSEE1312 pKa = 4.63SISEE1316 pKa = 4.19SQSLSISISEE1326 pKa = 4.26SDD1328 pKa = 3.63STSDD1332 pKa = 4.25SISDD1336 pKa = 3.98SEE1338 pKa = 4.52SDD1340 pKa = 4.25SISDD1344 pKa = 4.43SISDD1348 pKa = 4.46SISDD1352 pKa = 3.79STSDD1356 pKa = 4.02SISDD1360 pKa = 3.85SEE1362 pKa = 4.45SLSEE1366 pKa = 4.23SVSEE1370 pKa = 4.46SEE1372 pKa = 4.56SLSISISEE1380 pKa = 4.26SDD1382 pKa = 3.6STSDD1386 pKa = 3.51SLSDD1390 pKa = 3.91SEE1392 pKa = 5.25SDD1394 pKa = 3.94SISNSISDD1402 pKa = 4.81SISDD1406 pKa = 3.67STSNSISDD1414 pKa = 3.92SEE1416 pKa = 4.43SLSEE1420 pKa = 4.23SVSEE1424 pKa = 4.46SEE1426 pKa = 4.56SLSISISEE1434 pKa = 4.26SDD1436 pKa = 3.6STSDD1440 pKa = 3.51SLSDD1444 pKa = 3.91SEE1446 pKa = 4.94SDD1448 pKa = 4.24SISDD1452 pKa = 4.43SISDD1456 pKa = 4.51SISSSISNSDD1466 pKa = 3.5SLSEE1470 pKa = 4.16SVSKK1474 pKa = 11.08SEE1476 pKa = 4.38SLSISISEE1484 pKa = 4.33STSTSTSISDD1494 pKa = 3.58SSSTSTSTSNSDD1506 pKa = 3.24SDD1508 pKa = 4.02SLSNSISDD1516 pKa = 3.81SRR1518 pKa = 11.84SEE1520 pKa = 3.83
Molecular weight: 158.07 kDa
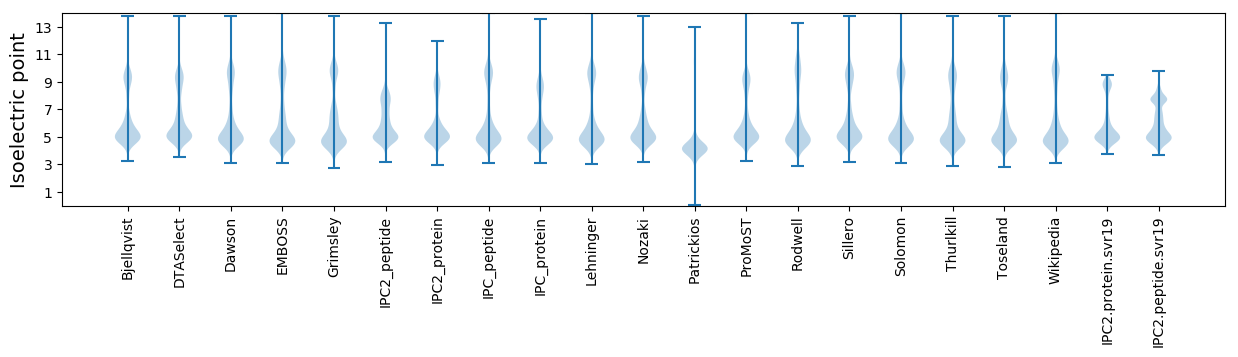
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R2QCP4|R2QCP4_9ENTE Alternate signal-mediated exported protein OS=Enterococcus pallens ATCC BAA-351 OX=1158607 GN=UAU_01942 PE=4 SV=1
RR1 pKa = 7.38IRR3 pKa = 11.84TVIQTRR9 pKa = 11.84TATQIPIATRR19 pKa = 11.84IRR21 pKa = 11.84TVIQILIVTRR31 pKa = 11.84IPIVIQTQTATQTRR45 pKa = 11.84TVIQTRR51 pKa = 11.84IVIQIRR57 pKa = 11.84TAIQIPIVTRR67 pKa = 11.84IPIVIRR73 pKa = 11.84IRR75 pKa = 11.84TVTQIPIATQTPIAIQILIRR95 pKa = 11.84NFRR98 pKa = 11.84QLL100 pKa = 3.1
RR1 pKa = 7.38IRR3 pKa = 11.84TVIQTRR9 pKa = 11.84TATQIPIATRR19 pKa = 11.84IRR21 pKa = 11.84TVIQILIVTRR31 pKa = 11.84IPIVIQTQTATQTRR45 pKa = 11.84TVIQTRR51 pKa = 11.84IVIQIRR57 pKa = 11.84TAIQIPIVTRR67 pKa = 11.84IPIVIRR73 pKa = 11.84IRR75 pKa = 11.84TVTQIPIATQTPIAIQILIRR95 pKa = 11.84NFRR98 pKa = 11.84QLL100 pKa = 3.1
Molecular weight: 11.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1566595 |
30 |
3111 |
302.4 |
33.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.327 ± 0.041 | 0.728 ± 0.011 |
5.251 ± 0.032 | 7.425 ± 0.044 |
4.478 ± 0.031 | 6.515 ± 0.037 |
1.74 ± 0.017 | 7.42 ± 0.039 |
6.583 ± 0.035 | 9.964 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.632 ± 0.019 | 4.555 ± 0.026 |
3.592 ± 0.02 | 4.486 ± 0.03 |
3.829 ± 0.024 | 6.143 ± 0.043 |
5.851 ± 0.038 | 6.809 ± 0.026 |
0.943 ± 0.012 | 3.73 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
