
Sewage-associated circular DNA virus-10
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.79
Get precalculated fractions of proteins
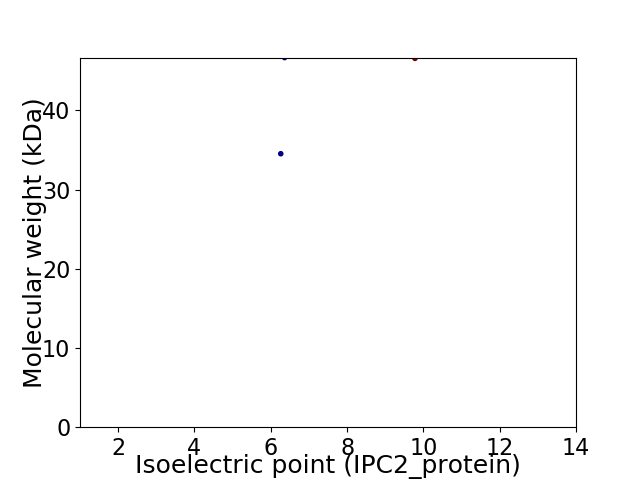
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A075J446|A0A075J446_9VIRU Capsid protein OS=Sewage-associated circular DNA virus-10 OX=1519386 PE=4 SV=1
MM1 pKa = 7.04SQNLEE6 pKa = 4.0SGRR9 pKa = 11.84RR10 pKa = 11.84EE11 pKa = 4.11ALKK14 pKa = 9.97MRR16 pKa = 11.84AWCFTHH22 pKa = 7.11NNFPYY27 pKa = 10.51AAADD31 pKa = 3.69LPLCKK36 pKa = 9.99DD37 pKa = 2.89EE38 pKa = 6.24RR39 pKa = 11.84YY40 pKa = 10.05VVWQHH45 pKa = 5.57EE46 pKa = 4.64KK47 pKa = 10.7VDD49 pKa = 3.61TDD51 pKa = 4.4HH52 pKa = 6.29IQGYY56 pKa = 9.76IEE58 pKa = 4.06LSKK61 pKa = 9.98PQRR64 pKa = 11.84ISAMIKK70 pKa = 8.6WLPGAHH76 pKa = 5.71FEE78 pKa = 4.35EE79 pKa = 5.13RR80 pKa = 11.84RR81 pKa = 11.84GTPDD85 pKa = 2.96QARR88 pKa = 11.84KK89 pKa = 10.14YY90 pKa = 10.79CMEE93 pKa = 4.25EE94 pKa = 3.82DD95 pKa = 3.74TRR97 pKa = 11.84VEE99 pKa = 4.47GPWEE103 pKa = 4.12RR104 pKa = 11.84GSYY107 pKa = 8.62GTTQGKK113 pKa = 10.06RR114 pKa = 11.84SDD116 pKa = 3.13IDD118 pKa = 3.61VVRR121 pKa = 11.84EE122 pKa = 4.38AIAAGADD129 pKa = 3.22RR130 pKa = 11.84RR131 pKa = 11.84EE132 pKa = 4.35VYY134 pKa = 9.84NAHH137 pKa = 6.6SDD139 pKa = 3.18IAAKK143 pKa = 9.81YY144 pKa = 7.76PRR146 pKa = 11.84YY147 pKa = 10.02VEE149 pKa = 3.95TMLRR153 pKa = 11.84FAKK156 pKa = 10.12EE157 pKa = 3.8DD158 pKa = 3.58AVDD161 pKa = 4.22KK162 pKa = 11.07ILVFEE167 pKa = 4.51PRR169 pKa = 11.84QGFQTDD175 pKa = 4.09LLDD178 pKa = 4.51MISGPADD185 pKa = 3.13SRR187 pKa = 11.84SIHH190 pKa = 5.14WVYY193 pKa = 11.47DD194 pKa = 3.47RR195 pKa = 11.84VGNNGKK201 pKa = 8.01TYY203 pKa = 9.04FAKK206 pKa = 10.91YY207 pKa = 10.3LVDD210 pKa = 3.82KK211 pKa = 11.1FGAFYY216 pKa = 10.56TNGGKK221 pKa = 9.79SVDD224 pKa = 3.27LAYY227 pKa = 10.25AYY229 pKa = 10.17GGEE232 pKa = 4.34SIVIFDD238 pKa = 3.74YY239 pKa = 11.36VRR241 pKa = 11.84DD242 pKa = 3.71AEE244 pKa = 4.4EE245 pKa = 3.86YY246 pKa = 9.18VGYY249 pKa = 10.63GVIEE253 pKa = 3.85QLKK256 pKa = 10.32NGIAMSTKK264 pKa = 9.9YY265 pKa = 10.88EE266 pKa = 4.59SITKK270 pKa = 9.99RR271 pKa = 11.84FNIPHH276 pKa = 6.34VIVLANFKK284 pKa = 9.51PQEE287 pKa = 4.27GKK289 pKa = 10.67FSSDD293 pKa = 3.55RR294 pKa = 11.84IKK296 pKa = 10.43MINVTT301 pKa = 3.77
MM1 pKa = 7.04SQNLEE6 pKa = 4.0SGRR9 pKa = 11.84RR10 pKa = 11.84EE11 pKa = 4.11ALKK14 pKa = 9.97MRR16 pKa = 11.84AWCFTHH22 pKa = 7.11NNFPYY27 pKa = 10.51AAADD31 pKa = 3.69LPLCKK36 pKa = 9.99DD37 pKa = 2.89EE38 pKa = 6.24RR39 pKa = 11.84YY40 pKa = 10.05VVWQHH45 pKa = 5.57EE46 pKa = 4.64KK47 pKa = 10.7VDD49 pKa = 3.61TDD51 pKa = 4.4HH52 pKa = 6.29IQGYY56 pKa = 9.76IEE58 pKa = 4.06LSKK61 pKa = 9.98PQRR64 pKa = 11.84ISAMIKK70 pKa = 8.6WLPGAHH76 pKa = 5.71FEE78 pKa = 4.35EE79 pKa = 5.13RR80 pKa = 11.84RR81 pKa = 11.84GTPDD85 pKa = 2.96QARR88 pKa = 11.84KK89 pKa = 10.14YY90 pKa = 10.79CMEE93 pKa = 4.25EE94 pKa = 3.82DD95 pKa = 3.74TRR97 pKa = 11.84VEE99 pKa = 4.47GPWEE103 pKa = 4.12RR104 pKa = 11.84GSYY107 pKa = 8.62GTTQGKK113 pKa = 10.06RR114 pKa = 11.84SDD116 pKa = 3.13IDD118 pKa = 3.61VVRR121 pKa = 11.84EE122 pKa = 4.38AIAAGADD129 pKa = 3.22RR130 pKa = 11.84RR131 pKa = 11.84EE132 pKa = 4.35VYY134 pKa = 9.84NAHH137 pKa = 6.6SDD139 pKa = 3.18IAAKK143 pKa = 9.81YY144 pKa = 7.76PRR146 pKa = 11.84YY147 pKa = 10.02VEE149 pKa = 3.95TMLRR153 pKa = 11.84FAKK156 pKa = 10.12EE157 pKa = 3.8DD158 pKa = 3.58AVDD161 pKa = 4.22KK162 pKa = 11.07ILVFEE167 pKa = 4.51PRR169 pKa = 11.84QGFQTDD175 pKa = 4.09LLDD178 pKa = 4.51MISGPADD185 pKa = 3.13SRR187 pKa = 11.84SIHH190 pKa = 5.14WVYY193 pKa = 11.47DD194 pKa = 3.47RR195 pKa = 11.84VGNNGKK201 pKa = 8.01TYY203 pKa = 9.04FAKK206 pKa = 10.91YY207 pKa = 10.3LVDD210 pKa = 3.82KK211 pKa = 11.1FGAFYY216 pKa = 10.56TNGGKK221 pKa = 9.79SVDD224 pKa = 3.27LAYY227 pKa = 10.25AYY229 pKa = 10.17GGEE232 pKa = 4.34SIVIFDD238 pKa = 3.74YY239 pKa = 11.36VRR241 pKa = 11.84DD242 pKa = 3.71AEE244 pKa = 4.4EE245 pKa = 3.86YY246 pKa = 9.18VGYY249 pKa = 10.63GVIEE253 pKa = 3.85QLKK256 pKa = 10.32NGIAMSTKK264 pKa = 9.9YY265 pKa = 10.88EE266 pKa = 4.59SITKK270 pKa = 9.99RR271 pKa = 11.84FNIPHH276 pKa = 6.34VIVLANFKK284 pKa = 9.51PQEE287 pKa = 4.27GKK289 pKa = 10.67FSSDD293 pKa = 3.55RR294 pKa = 11.84IKK296 pKa = 10.43MINVTT301 pKa = 3.77
Molecular weight: 34.54 kDa
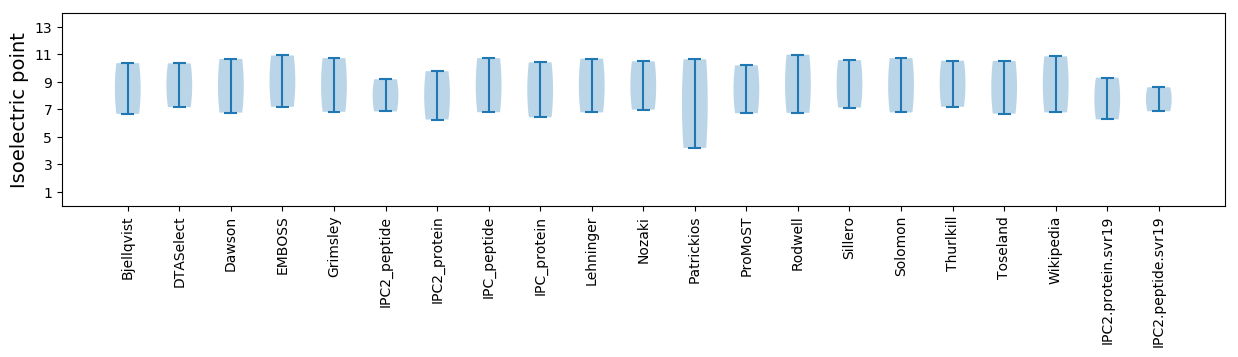
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A075J446|A0A075J446_9VIRU Capsid protein OS=Sewage-associated circular DNA virus-10 OX=1519386 PE=4 SV=1
MM1 pKa = 6.67YY2 pKa = 10.15RR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84KK6 pKa = 6.37TARR9 pKa = 11.84KK10 pKa = 8.21SRR12 pKa = 11.84RR13 pKa = 11.84VKK15 pKa = 9.48TRR17 pKa = 11.84RR18 pKa = 11.84SSLGRR23 pKa = 11.84RR24 pKa = 11.84KK25 pKa = 9.85RR26 pKa = 11.84SLRR29 pKa = 11.84KK30 pKa = 8.5VSRR33 pKa = 11.84RR34 pKa = 11.84KK35 pKa = 9.8RR36 pKa = 11.84KK37 pKa = 7.64STPRR41 pKa = 11.84SGMKK45 pKa = 9.14MRR47 pKa = 11.84RR48 pKa = 11.84SVAKK52 pKa = 10.01SANPTMMDD60 pKa = 3.29VQAVAIEE67 pKa = 4.06TSKK70 pKa = 10.83RR71 pKa = 11.84SSARR75 pKa = 11.84LKK77 pKa = 10.91SRR79 pKa = 11.84GNFINKK85 pKa = 8.51VVTANLQSSILRR97 pKa = 11.84FQAINRR103 pKa = 11.84YY104 pKa = 6.85MNRR107 pKa = 11.84TEE109 pKa = 4.07LQRR112 pKa = 11.84NTAFTYY118 pKa = 10.28GGAYY122 pKa = 10.16SLQNVGVPTQTFCPLHH138 pKa = 7.5LYY140 pKa = 10.58DD141 pKa = 4.38ITCSPNIVNTTVTQPVVAQVLTLRR165 pKa = 11.84TGAVPFSTPTSTNFEE180 pKa = 4.33YY181 pKa = 10.69LYY183 pKa = 10.9GDD185 pKa = 3.89SLAGSDD191 pKa = 4.17GVSTGWQQEE200 pKa = 4.02QLPSGFGTVVYY211 pKa = 9.68PNRR214 pKa = 11.84RR215 pKa = 11.84TLLEE219 pKa = 4.2WVSLDD224 pKa = 3.37MFCYY228 pKa = 10.0GAILMPVTYY237 pKa = 8.93TVSVVKK243 pKa = 10.36FSKK246 pKa = 10.5DD247 pKa = 3.37YY248 pKa = 10.62LHH250 pKa = 7.25PGMGGIDD257 pKa = 3.75FNAIDD262 pKa = 4.66DD263 pKa = 4.04VRR265 pKa = 11.84LDD267 pKa = 4.38RR268 pKa = 11.84LNFWLRR274 pKa = 11.84QIKK277 pKa = 10.34SDD279 pKa = 3.21ISHH282 pKa = 7.33PLALEE287 pKa = 4.26GQTGNARR294 pKa = 11.84LKK296 pKa = 10.21VLKK299 pKa = 8.62TQSFTLQPSSNAVGEE314 pKa = 4.29GGITNPAPRR323 pKa = 11.84TKK325 pKa = 10.47RR326 pKa = 11.84FTMKK330 pKa = 10.35IAINDD335 pKa = 3.41VMKK338 pKa = 10.87YY339 pKa = 10.26DD340 pKa = 3.19WNDD343 pKa = 3.26RR344 pKa = 11.84VNTDD348 pKa = 4.29NNILDD353 pKa = 4.24PGSTIEE359 pKa = 4.53NKK361 pKa = 10.8GEE363 pKa = 3.53MSTYY367 pKa = 9.98VDD369 pKa = 3.61YY370 pKa = 11.19KK371 pKa = 9.8KK372 pKa = 10.39RR373 pKa = 11.84IYY375 pKa = 10.84IMVQATNRR383 pKa = 11.84APTEE387 pKa = 3.99NDD389 pKa = 2.9SSDD392 pKa = 2.77WRR394 pKa = 11.84FSPSYY399 pKa = 10.62DD400 pKa = 2.74IVIRR404 pKa = 11.84KK405 pKa = 9.54KK406 pKa = 9.48YY407 pKa = 9.26TNITT411 pKa = 3.44
MM1 pKa = 6.67YY2 pKa = 10.15RR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84KK6 pKa = 6.37TARR9 pKa = 11.84KK10 pKa = 8.21SRR12 pKa = 11.84RR13 pKa = 11.84VKK15 pKa = 9.48TRR17 pKa = 11.84RR18 pKa = 11.84SSLGRR23 pKa = 11.84RR24 pKa = 11.84KK25 pKa = 9.85RR26 pKa = 11.84SLRR29 pKa = 11.84KK30 pKa = 8.5VSRR33 pKa = 11.84RR34 pKa = 11.84KK35 pKa = 9.8RR36 pKa = 11.84KK37 pKa = 7.64STPRR41 pKa = 11.84SGMKK45 pKa = 9.14MRR47 pKa = 11.84RR48 pKa = 11.84SVAKK52 pKa = 10.01SANPTMMDD60 pKa = 3.29VQAVAIEE67 pKa = 4.06TSKK70 pKa = 10.83RR71 pKa = 11.84SSARR75 pKa = 11.84LKK77 pKa = 10.91SRR79 pKa = 11.84GNFINKK85 pKa = 8.51VVTANLQSSILRR97 pKa = 11.84FQAINRR103 pKa = 11.84YY104 pKa = 6.85MNRR107 pKa = 11.84TEE109 pKa = 4.07LQRR112 pKa = 11.84NTAFTYY118 pKa = 10.28GGAYY122 pKa = 10.16SLQNVGVPTQTFCPLHH138 pKa = 7.5LYY140 pKa = 10.58DD141 pKa = 4.38ITCSPNIVNTTVTQPVVAQVLTLRR165 pKa = 11.84TGAVPFSTPTSTNFEE180 pKa = 4.33YY181 pKa = 10.69LYY183 pKa = 10.9GDD185 pKa = 3.89SLAGSDD191 pKa = 4.17GVSTGWQQEE200 pKa = 4.02QLPSGFGTVVYY211 pKa = 9.68PNRR214 pKa = 11.84RR215 pKa = 11.84TLLEE219 pKa = 4.2WVSLDD224 pKa = 3.37MFCYY228 pKa = 10.0GAILMPVTYY237 pKa = 8.93TVSVVKK243 pKa = 10.36FSKK246 pKa = 10.5DD247 pKa = 3.37YY248 pKa = 10.62LHH250 pKa = 7.25PGMGGIDD257 pKa = 3.75FNAIDD262 pKa = 4.66DD263 pKa = 4.04VRR265 pKa = 11.84LDD267 pKa = 4.38RR268 pKa = 11.84LNFWLRR274 pKa = 11.84QIKK277 pKa = 10.34SDD279 pKa = 3.21ISHH282 pKa = 7.33PLALEE287 pKa = 4.26GQTGNARR294 pKa = 11.84LKK296 pKa = 10.21VLKK299 pKa = 8.62TQSFTLQPSSNAVGEE314 pKa = 4.29GGITNPAPRR323 pKa = 11.84TKK325 pKa = 10.47RR326 pKa = 11.84FTMKK330 pKa = 10.35IAINDD335 pKa = 3.41VMKK338 pKa = 10.87YY339 pKa = 10.26DD340 pKa = 3.19WNDD343 pKa = 3.26RR344 pKa = 11.84VNTDD348 pKa = 4.29NNILDD353 pKa = 4.24PGSTIEE359 pKa = 4.53NKK361 pKa = 10.8GEE363 pKa = 3.53MSTYY367 pKa = 9.98VDD369 pKa = 3.61YY370 pKa = 11.19KK371 pKa = 9.8KK372 pKa = 10.39RR373 pKa = 11.84IYY375 pKa = 10.84IMVQATNRR383 pKa = 11.84APTEE387 pKa = 3.99NDD389 pKa = 2.9SSDD392 pKa = 2.77WRR394 pKa = 11.84FSPSYY399 pKa = 10.62DD400 pKa = 2.74IVIRR404 pKa = 11.84KK405 pKa = 9.54KK406 pKa = 9.48YY407 pKa = 9.26TNITT411 pKa = 3.44
Molecular weight: 46.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
712 |
301 |
411 |
356.0 |
40.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.601 ± 0.985 | 0.843 ± 0.089 |
5.758 ± 0.704 | 4.494 ± 1.626 |
3.792 ± 0.304 | 6.461 ± 0.49 |
1.404 ± 0.532 | 5.618 ± 0.401 |
6.32 ± 0.187 | 5.899 ± 0.721 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.949 ± 0.168 | 5.197 ± 0.891 |
4.213 ± 0.323 | 3.652 ± 0.19 |
8.567 ± 0.727 | 7.444 ± 1.23 |
7.163 ± 1.643 | 7.303 ± 0.189 |
1.404 ± 0.148 | 4.916 ± 0.615 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
