
Cellulosimicrobium sp. CUA-896
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Promicromonosporaceae; Cellulosimicrobium; unclassified Cellulosimicrobium
Average proteome isoelectric point is 6.27
Get precalculated fractions of proteins
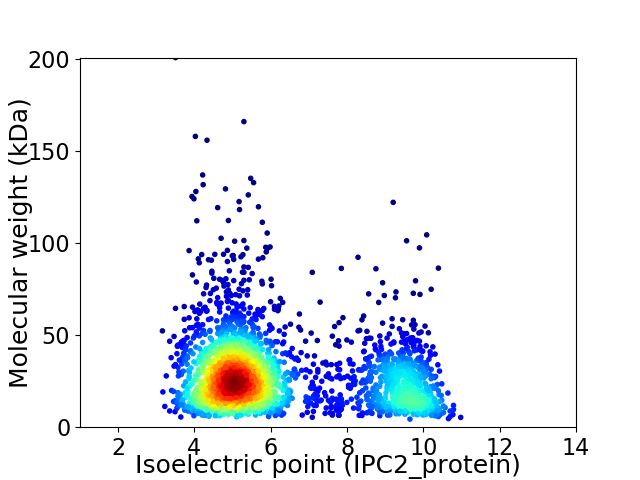
Virtual 2D-PAGE plot for 2595 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Q9VTY0|A0A1Q9VTY0_9MICO 4-hydroxy-3-methylbut-2-enyl diphosphate reductase OS=Cellulosimicrobium sp. CUA-896 OX=1517881 GN=ispH PE=3 SV=1
MM1 pKa = 7.8RR2 pKa = 11.84NRR4 pKa = 11.84RR5 pKa = 11.84FAALTALGLSGVLALTACSSGSDD28 pKa = 4.77DD29 pKa = 4.47DD30 pKa = 5.34AQDD33 pKa = 3.62ASSSGSSTGVVTVSNGEE50 pKa = 4.02PQNPLIPTMTNEE62 pKa = 4.47TNGSLTVQNIFSGLVYY78 pKa = 10.46YY79 pKa = 10.48DD80 pKa = 3.87AEE82 pKa = 4.79GGVHH86 pKa = 6.27NEE88 pKa = 3.69VAEE91 pKa = 4.56SIEE94 pKa = 4.88SEE96 pKa = 4.88DD97 pKa = 3.58NQAWTITIADD107 pKa = 3.62GWTFTDD113 pKa = 4.29GTPVTAEE120 pKa = 4.16SFASAWDD127 pKa = 3.45YY128 pKa = 11.6GAALDD133 pKa = 3.96NAQNLSYY140 pKa = 10.31FFEE143 pKa = 5.52PIQGFSYY150 pKa = 10.79DD151 pKa = 3.88ANTSLIEE158 pKa = 4.06EE159 pKa = 4.76GGLVVEE165 pKa = 5.21DD166 pKa = 4.6EE167 pKa = 4.5STLVVNLTQPEE178 pKa = 4.24SDD180 pKa = 3.48FPIRR184 pKa = 11.84LGYY187 pKa = 10.46AAFFPLPEE195 pKa = 4.7AFFDD199 pKa = 3.98DD200 pKa = 4.42PEE202 pKa = 5.75AFGEE206 pKa = 4.29APVGNGPYY214 pKa = 9.51MLEE217 pKa = 3.98TWEE220 pKa = 4.41HH221 pKa = 6.9DD222 pKa = 3.68SLISLVPNPDD232 pKa = 3.2YY233 pKa = 11.26AGEE236 pKa = 4.0RR237 pKa = 11.84TPANGGLEE245 pKa = 4.4LIAYY249 pKa = 7.76TDD251 pKa = 3.52EE252 pKa = 4.53DD253 pKa = 3.65SAYY256 pKa = 10.87NDD258 pKa = 3.97LLGGQLDD265 pKa = 4.02IIKK268 pKa = 9.85NVPSSAFATFEE279 pKa = 4.38DD280 pKa = 4.58EE281 pKa = 4.79LEE283 pKa = 4.34GRR285 pKa = 11.84AVNQPAAVIQTLTVPEE301 pKa = 4.52WLPEE305 pKa = 4.09FQGEE309 pKa = 4.11AGLLRR314 pKa = 11.84RR315 pKa = 11.84QAISHH320 pKa = 6.88AIDD323 pKa = 3.42RR324 pKa = 11.84DD325 pKa = 3.9EE326 pKa = 4.0VTEE329 pKa = 4.48TIFAGSRR336 pKa = 11.84TPATDD341 pKa = 3.39FTSPVIDD348 pKa = 3.43GWTEE352 pKa = 3.93EE353 pKa = 4.07IPGNEE358 pKa = 3.94VLEE361 pKa = 4.18YY362 pKa = 10.84DD363 pKa = 3.82ADD365 pKa = 4.0LAAEE369 pKa = 4.42LWGEE373 pKa = 4.16AEE375 pKa = 4.59AIEE378 pKa = 4.26PVGDD382 pKa = 3.55YY383 pKa = 9.92TLQIASNADD392 pKa = 3.24SDD394 pKa = 3.84HH395 pKa = 5.38QTWVDD400 pKa = 3.57AVCNNIRR407 pKa = 11.84TVLEE411 pKa = 3.96IGCEE415 pKa = 3.95FFPYY419 pKa = 8.83PTFDD423 pKa = 3.43EE424 pKa = 4.44YY425 pKa = 11.77LDD427 pKa = 4.11ARR429 pKa = 11.84DD430 pKa = 3.95SGSVPGPFRR439 pKa = 11.84GGWQADD445 pKa = 3.86YY446 pKa = 9.61PAMSNFLGPIYY457 pKa = 9.96GTGAGSNDD465 pKa = 4.04GQWSNADD472 pKa = 3.07FDD474 pKa = 4.24ATLTEE479 pKa = 4.47ANSAEE484 pKa = 4.15TRR486 pKa = 11.84SRR488 pKa = 11.84RR489 pKa = 11.84SSCTWTPRR497 pKa = 11.84RR498 pKa = 11.84SCSRR502 pKa = 11.84TCRR505 pKa = 11.84ALPLWYY511 pKa = 10.69SNATGGWADD520 pKa = 3.41TVEE523 pKa = 4.0NVEE526 pKa = 4.84FGWDD530 pKa = 3.68SDD532 pKa = 4.44PILYY536 pKa = 10.08QVTKK540 pKa = 11.13AEE542 pKa = 4.09
MM1 pKa = 7.8RR2 pKa = 11.84NRR4 pKa = 11.84RR5 pKa = 11.84FAALTALGLSGVLALTACSSGSDD28 pKa = 4.77DD29 pKa = 4.47DD30 pKa = 5.34AQDD33 pKa = 3.62ASSSGSSTGVVTVSNGEE50 pKa = 4.02PQNPLIPTMTNEE62 pKa = 4.47TNGSLTVQNIFSGLVYY78 pKa = 10.46YY79 pKa = 10.48DD80 pKa = 3.87AEE82 pKa = 4.79GGVHH86 pKa = 6.27NEE88 pKa = 3.69VAEE91 pKa = 4.56SIEE94 pKa = 4.88SEE96 pKa = 4.88DD97 pKa = 3.58NQAWTITIADD107 pKa = 3.62GWTFTDD113 pKa = 4.29GTPVTAEE120 pKa = 4.16SFASAWDD127 pKa = 3.45YY128 pKa = 11.6GAALDD133 pKa = 3.96NAQNLSYY140 pKa = 10.31FFEE143 pKa = 5.52PIQGFSYY150 pKa = 10.79DD151 pKa = 3.88ANTSLIEE158 pKa = 4.06EE159 pKa = 4.76GGLVVEE165 pKa = 5.21DD166 pKa = 4.6EE167 pKa = 4.5STLVVNLTQPEE178 pKa = 4.24SDD180 pKa = 3.48FPIRR184 pKa = 11.84LGYY187 pKa = 10.46AAFFPLPEE195 pKa = 4.7AFFDD199 pKa = 3.98DD200 pKa = 4.42PEE202 pKa = 5.75AFGEE206 pKa = 4.29APVGNGPYY214 pKa = 9.51MLEE217 pKa = 3.98TWEE220 pKa = 4.41HH221 pKa = 6.9DD222 pKa = 3.68SLISLVPNPDD232 pKa = 3.2YY233 pKa = 11.26AGEE236 pKa = 4.0RR237 pKa = 11.84TPANGGLEE245 pKa = 4.4LIAYY249 pKa = 7.76TDD251 pKa = 3.52EE252 pKa = 4.53DD253 pKa = 3.65SAYY256 pKa = 10.87NDD258 pKa = 3.97LLGGQLDD265 pKa = 4.02IIKK268 pKa = 9.85NVPSSAFATFEE279 pKa = 4.38DD280 pKa = 4.58EE281 pKa = 4.79LEE283 pKa = 4.34GRR285 pKa = 11.84AVNQPAAVIQTLTVPEE301 pKa = 4.52WLPEE305 pKa = 4.09FQGEE309 pKa = 4.11AGLLRR314 pKa = 11.84RR315 pKa = 11.84QAISHH320 pKa = 6.88AIDD323 pKa = 3.42RR324 pKa = 11.84DD325 pKa = 3.9EE326 pKa = 4.0VTEE329 pKa = 4.48TIFAGSRR336 pKa = 11.84TPATDD341 pKa = 3.39FTSPVIDD348 pKa = 3.43GWTEE352 pKa = 3.93EE353 pKa = 4.07IPGNEE358 pKa = 3.94VLEE361 pKa = 4.18YY362 pKa = 10.84DD363 pKa = 3.82ADD365 pKa = 4.0LAAEE369 pKa = 4.42LWGEE373 pKa = 4.16AEE375 pKa = 4.59AIEE378 pKa = 4.26PVGDD382 pKa = 3.55YY383 pKa = 9.92TLQIASNADD392 pKa = 3.24SDD394 pKa = 3.84HH395 pKa = 5.38QTWVDD400 pKa = 3.57AVCNNIRR407 pKa = 11.84TVLEE411 pKa = 3.96IGCEE415 pKa = 3.95FFPYY419 pKa = 8.83PTFDD423 pKa = 3.43EE424 pKa = 4.44YY425 pKa = 11.77LDD427 pKa = 4.11ARR429 pKa = 11.84DD430 pKa = 3.95SGSVPGPFRR439 pKa = 11.84GGWQADD445 pKa = 3.86YY446 pKa = 9.61PAMSNFLGPIYY457 pKa = 9.96GTGAGSNDD465 pKa = 4.04GQWSNADD472 pKa = 3.07FDD474 pKa = 4.24ATLTEE479 pKa = 4.47ANSAEE484 pKa = 4.15TRR486 pKa = 11.84SRR488 pKa = 11.84RR489 pKa = 11.84SSCTWTPRR497 pKa = 11.84RR498 pKa = 11.84SCSRR502 pKa = 11.84TCRR505 pKa = 11.84ALPLWYY511 pKa = 10.69SNATGGWADD520 pKa = 3.41TVEE523 pKa = 4.0NVEE526 pKa = 4.84FGWDD530 pKa = 3.68SDD532 pKa = 4.44PILYY536 pKa = 10.08QVTKK540 pKa = 11.13AEE542 pKa = 4.09
Molecular weight: 58.64 kDa
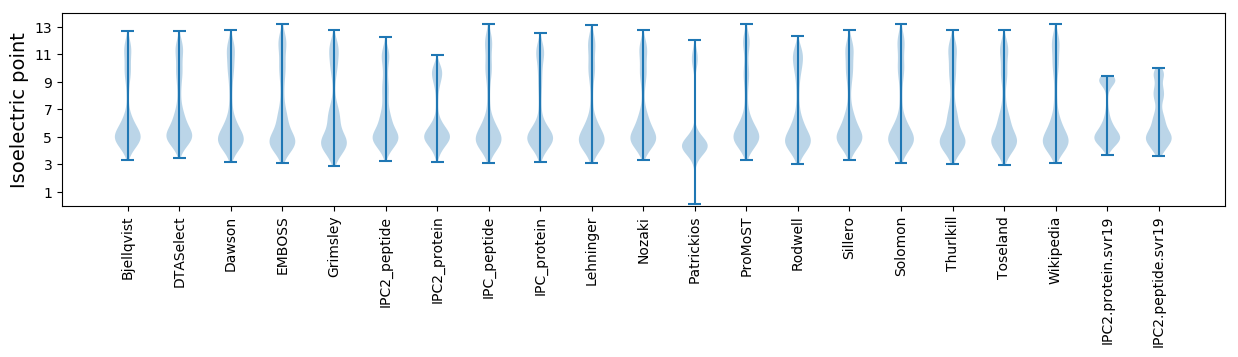
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Q9VWY3|A0A1Q9VWY3_9MICO Helicase ATP-binding domain-containing protein OS=Cellulosimicrobium sp. CUA-896 OX=1517881 GN=BJF88_09305 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 8.98RR4 pKa = 11.84TFQPNTRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.83THH17 pKa = 5.18GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.97GRR40 pKa = 11.84TEE42 pKa = 3.99LSAA45 pKa = 4.86
MM1 pKa = 7.69SKK3 pKa = 8.98RR4 pKa = 11.84TFQPNTRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.83THH17 pKa = 5.18GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.97GRR40 pKa = 11.84TEE42 pKa = 3.99LSAA45 pKa = 4.86
Molecular weight: 5.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
732398 |
37 |
1957 |
282.2 |
29.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.639 ± 0.079 | 0.623 ± 0.013 |
6.596 ± 0.052 | 5.442 ± 0.051 |
2.396 ± 0.032 | 9.608 ± 0.049 |
2.009 ± 0.025 | 2.403 ± 0.037 |
1.19 ± 0.027 | 9.852 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.471 ± 0.019 | 1.332 ± 0.026 |
6.193 ± 0.047 | 2.395 ± 0.028 |
8.453 ± 0.069 | 5.214 ± 0.045 |
6.456 ± 0.048 | 10.491 ± 0.055 |
1.514 ± 0.021 | 1.723 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
