
Thermodesulfatator indicus (strain DSM 15286 / JCM 11887 / CIR29812)
Taxonomy: cellular organisms; Bacteria; Thermodesulfobacteria; Thermodesulfobacteria; Thermodesulfobacteriales; Thermodesulfobacteriaceae; Thermodesulfatator; Thermodesulfatator indicus
Average proteome isoelectric point is 7.16
Get precalculated fractions of proteins
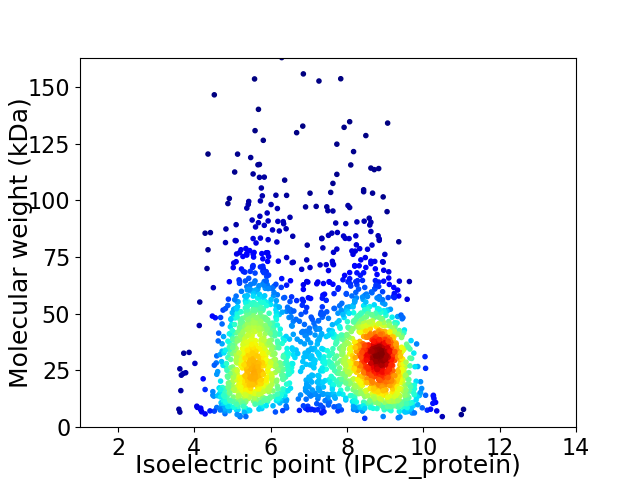
Virtual 2D-PAGE plot for 2184 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
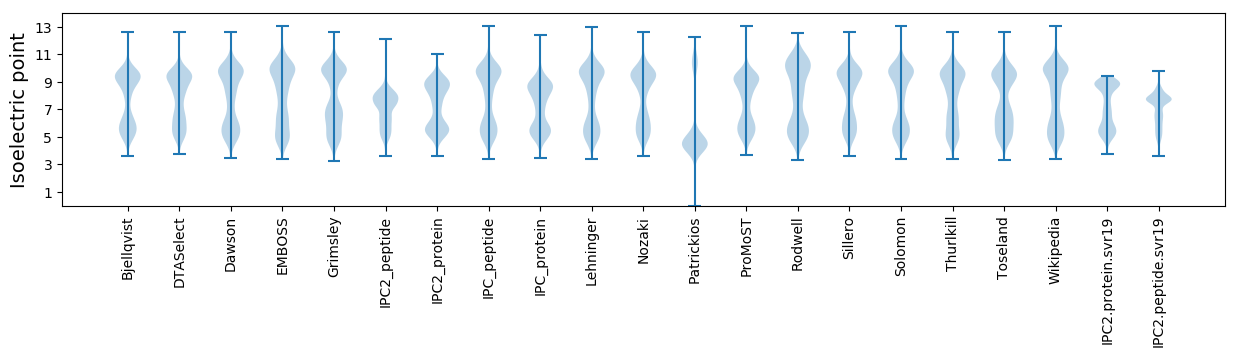
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F8AAB1|F8AAB1_THEID Primosomal protein N' OS=Thermodesulfatator indicus (strain DSM 15286 / JCM 11887 / CIR29812) OX=667014 GN=priA PE=3 SV=1
MM1 pKa = 7.26FKK3 pKa = 10.49KK4 pKa = 10.59VLSSLFLAVLMLGLAVSNAKK24 pKa = 9.67AAPIVMEE31 pKa = 4.02GNYY34 pKa = 10.1VYY36 pKa = 10.01TQVSEE41 pKa = 4.96DD42 pKa = 3.43GTLGNGFNFPGIQYY56 pKa = 9.88DD57 pKa = 3.78PSGTASFPGSGGKK70 pKa = 9.96DD71 pKa = 3.19FLQPGDD77 pKa = 3.97PFEE80 pKa = 4.64GFYY83 pKa = 11.0LKK85 pKa = 10.86SNEE88 pKa = 4.28SGIIGNNNDD97 pKa = 2.53WSVFIGTLTDD107 pKa = 4.07LSGSGYY113 pKa = 10.81DD114 pKa = 4.44NFIRR118 pKa = 11.84WEE120 pKa = 4.47GSLGTYY126 pKa = 9.98FDD128 pKa = 4.11VAIEE132 pKa = 4.53TYY134 pKa = 10.75FNDD137 pKa = 2.89SDD139 pKa = 4.16KK140 pKa = 11.03FIKK143 pKa = 10.67FSTTVTALNDD153 pKa = 3.41LTDD156 pKa = 3.46VYY158 pKa = 10.19FLRR161 pKa = 11.84VIDD164 pKa = 4.88PDD166 pKa = 3.15QDD168 pKa = 3.82TNDD171 pKa = 3.43FGIFDD176 pKa = 4.22TNNARR181 pKa = 11.84GFGSFSPEE189 pKa = 3.48DD190 pKa = 3.25WVYY193 pKa = 11.17AAGPISNWTIGLYY206 pKa = 10.13SDD208 pKa = 4.59SLVPHH213 pKa = 5.7NTGVSFAWSGDD224 pKa = 3.1PEE226 pKa = 4.44FYY228 pKa = 11.21YY229 pKa = 11.02NGNNDD234 pKa = 3.08GNGDD238 pKa = 3.79YY239 pKa = 10.86TIGLAFYY246 pKa = 10.67LGDD249 pKa = 3.88LSVGDD254 pKa = 4.41SVTFDD259 pKa = 3.33YY260 pKa = 11.18YY261 pKa = 11.48YY262 pKa = 11.64VLGTSPADD270 pKa = 3.2AASNIPVASPIPEE283 pKa = 4.53PSTVVLIGVGLGALGFYY300 pKa = 10.46RR301 pKa = 11.84SRR303 pKa = 11.84RR304 pKa = 11.84KK305 pKa = 9.76
MM1 pKa = 7.26FKK3 pKa = 10.49KK4 pKa = 10.59VLSSLFLAVLMLGLAVSNAKK24 pKa = 9.67AAPIVMEE31 pKa = 4.02GNYY34 pKa = 10.1VYY36 pKa = 10.01TQVSEE41 pKa = 4.96DD42 pKa = 3.43GTLGNGFNFPGIQYY56 pKa = 9.88DD57 pKa = 3.78PSGTASFPGSGGKK70 pKa = 9.96DD71 pKa = 3.19FLQPGDD77 pKa = 3.97PFEE80 pKa = 4.64GFYY83 pKa = 11.0LKK85 pKa = 10.86SNEE88 pKa = 4.28SGIIGNNNDD97 pKa = 2.53WSVFIGTLTDD107 pKa = 4.07LSGSGYY113 pKa = 10.81DD114 pKa = 4.44NFIRR118 pKa = 11.84WEE120 pKa = 4.47GSLGTYY126 pKa = 9.98FDD128 pKa = 4.11VAIEE132 pKa = 4.53TYY134 pKa = 10.75FNDD137 pKa = 2.89SDD139 pKa = 4.16KK140 pKa = 11.03FIKK143 pKa = 10.67FSTTVTALNDD153 pKa = 3.41LTDD156 pKa = 3.46VYY158 pKa = 10.19FLRR161 pKa = 11.84VIDD164 pKa = 4.88PDD166 pKa = 3.15QDD168 pKa = 3.82TNDD171 pKa = 3.43FGIFDD176 pKa = 4.22TNNARR181 pKa = 11.84GFGSFSPEE189 pKa = 3.48DD190 pKa = 3.25WVYY193 pKa = 11.17AAGPISNWTIGLYY206 pKa = 10.13SDD208 pKa = 4.59SLVPHH213 pKa = 5.7NTGVSFAWSGDD224 pKa = 3.1PEE226 pKa = 4.44FYY228 pKa = 11.21YY229 pKa = 11.02NGNNDD234 pKa = 3.08GNGDD238 pKa = 3.79YY239 pKa = 10.86TIGLAFYY246 pKa = 10.67LGDD249 pKa = 3.88LSVGDD254 pKa = 4.41SVTFDD259 pKa = 3.33YY260 pKa = 11.18YY261 pKa = 11.48YY262 pKa = 11.64VLGTSPADD270 pKa = 3.2AASNIPVASPIPEE283 pKa = 4.53PSTVVLIGVGLGALGFYY300 pKa = 10.46RR301 pKa = 11.84SRR303 pKa = 11.84RR304 pKa = 11.84KK305 pKa = 9.76
Molecular weight: 32.97 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F8ABU2|F8ABU2_THEID Cysteine--tRNA ligase OS=Thermodesulfatator indicus (strain DSM 15286 / JCM 11887 / CIR29812) OX=667014 GN=cysS PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 9.91QPSRR9 pKa = 11.84IKK11 pKa = 10.5RR12 pKa = 11.84KK13 pKa = 8.27RR14 pKa = 11.84CHH16 pKa = 5.7GFRR19 pKa = 11.84ARR21 pKa = 11.84MRR23 pKa = 11.84TRR25 pKa = 11.84SGRR28 pKa = 11.84RR29 pKa = 11.84ILANRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.74GRR39 pKa = 11.84WRR41 pKa = 11.84LTVV44 pKa = 3.0
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 9.91QPSRR9 pKa = 11.84IKK11 pKa = 10.5RR12 pKa = 11.84KK13 pKa = 8.27RR14 pKa = 11.84CHH16 pKa = 5.7GFRR19 pKa = 11.84ARR21 pKa = 11.84MRR23 pKa = 11.84TRR25 pKa = 11.84SGRR28 pKa = 11.84RR29 pKa = 11.84ILANRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.74GRR39 pKa = 11.84WRR41 pKa = 11.84LTVV44 pKa = 3.0
Molecular weight: 5.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
694740 |
35 |
1408 |
318.1 |
35.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.625 ± 0.059 | 1.152 ± 0.025 |
4.362 ± 0.031 | 8.21 ± 0.067 |
5.003 ± 0.044 | 6.765 ± 0.044 |
1.811 ± 0.022 | 7.25 ± 0.049 |
7.993 ± 0.059 | 11.425 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.831 ± 0.018 | 3.277 ± 0.039 |
4.812 ± 0.032 | 2.811 ± 0.029 |
5.345 ± 0.042 | 4.953 ± 0.036 |
4.299 ± 0.033 | 6.707 ± 0.043 |
1.098 ± 0.02 | 3.272 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
