
Salpingoeca rosetta (strain ATCC 50818 / BSB-021)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Choanoflagellata; Craspedida; Salpingoecidae; Salpingoeca
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
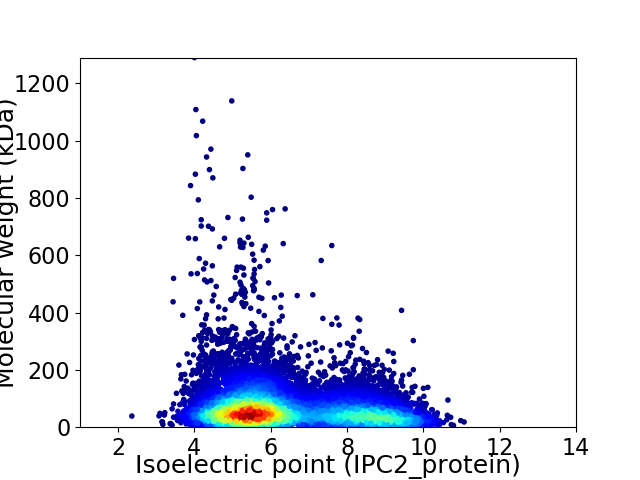
Virtual 2D-PAGE plot for 11698 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|F2U247|F2U247_SALR5 Uncharacterized protein OS=Salpingoeca rosetta (strain ATCC 50818 / BSB-021) OX=946362 GN=PTSG_02412 PE=4 SV=1
MM1 pKa = 7.75KK2 pKa = 9.56LTFALVAAAAILACATAQDD21 pKa = 3.99IFGVWTGPVSPGSDD35 pKa = 3.16CSILGEE41 pKa = 3.97FRR43 pKa = 11.84EE44 pKa = 4.53TSFFTIISGEE54 pKa = 4.08EE55 pKa = 3.83SSGCASGIFIGSLTVTSPASGSSADD80 pKa = 3.42TSIIITYY87 pKa = 9.92VEE89 pKa = 4.37VPDD92 pKa = 4.57FATDD96 pKa = 3.51QISPGDD102 pKa = 3.56TFVFDD107 pKa = 4.65IYY109 pKa = 11.15LSGDD113 pKa = 3.22TMTLRR118 pKa = 11.84NDD120 pKa = 3.37YY121 pKa = 11.1FEE123 pKa = 4.68TQGLWPTGGLVLTTYY138 pKa = 9.1PAWPINGYY146 pKa = 9.13WDD148 pKa = 4.23GNDD151 pKa = 3.24TSLLYY156 pKa = 10.46INNGLYY162 pKa = 10.57LRR164 pKa = 11.84ASAFGEE170 pKa = 4.3GGEE173 pKa = 4.34GSSAGVVEE181 pKa = 4.39MTGNTDD187 pKa = 2.86EE188 pKa = 4.18YY189 pKa = 9.52MSRR192 pKa = 11.84IVGSVSDD199 pKa = 4.4IEE201 pKa = 4.95GDD203 pKa = 3.53TYY205 pKa = 11.19TFTMPSSGDD214 pKa = 3.13TRR216 pKa = 11.84LLDD219 pKa = 4.17FSDD222 pKa = 4.05PADD225 pKa = 3.43MNGFLLTKK233 pKa = 10.73NEE235 pKa = 4.02DD236 pKa = 3.53DD237 pKa = 4.21EE238 pKa = 5.62FNGIYY243 pKa = 10.44YY244 pKa = 10.2GVSPAQGEE252 pKa = 4.53DD253 pKa = 3.55CMQSTDD259 pKa = 3.83FNNGIFRR266 pKa = 11.84AFRR269 pKa = 11.84FQCDD273 pKa = 3.69DD274 pKa = 4.16SISDD278 pKa = 3.55DD279 pKa = 3.65GLRR282 pKa = 11.84VSAYY286 pKa = 8.27IGRR289 pKa = 11.84YY290 pKa = 5.28TVSGNQMRR298 pKa = 11.84LLYY301 pKa = 10.07TYY303 pKa = 9.98AAPDD307 pKa = 3.95DD308 pKa = 4.58DD309 pKa = 4.58LAGAEE314 pKa = 4.44VVFTYY319 pKa = 9.71TRR321 pKa = 11.84SGRR324 pKa = 11.84QLTLTGSVGGSDD336 pKa = 3.51EE337 pKa = 4.17FTITMNRR344 pKa = 11.84ADD346 pKa = 4.72RR347 pKa = 11.84APAATVVNITVSGDD361 pKa = 3.31VNTFDD366 pKa = 5.44ADD368 pKa = 3.31AFLAGLAQVLDD379 pKa = 3.7IPVNTIEE386 pKa = 4.54VVSISPGSVVVVVAIRR402 pKa = 11.84EE403 pKa = 4.15DD404 pKa = 3.81ASDD407 pKa = 3.39SSAPSLSTIIKK418 pKa = 10.03RR419 pKa = 11.84IEE421 pKa = 3.95AVPAGTSIGGYY432 pKa = 8.85GLEE435 pKa = 4.2SAQASRR441 pKa = 11.84DD442 pKa = 3.82PGSTSAASVATTSWFAAAILFMAVVSQLLAA472 pKa = 4.53
MM1 pKa = 7.75KK2 pKa = 9.56LTFALVAAAAILACATAQDD21 pKa = 3.99IFGVWTGPVSPGSDD35 pKa = 3.16CSILGEE41 pKa = 3.97FRR43 pKa = 11.84EE44 pKa = 4.53TSFFTIISGEE54 pKa = 4.08EE55 pKa = 3.83SSGCASGIFIGSLTVTSPASGSSADD80 pKa = 3.42TSIIITYY87 pKa = 9.92VEE89 pKa = 4.37VPDD92 pKa = 4.57FATDD96 pKa = 3.51QISPGDD102 pKa = 3.56TFVFDD107 pKa = 4.65IYY109 pKa = 11.15LSGDD113 pKa = 3.22TMTLRR118 pKa = 11.84NDD120 pKa = 3.37YY121 pKa = 11.1FEE123 pKa = 4.68TQGLWPTGGLVLTTYY138 pKa = 9.1PAWPINGYY146 pKa = 9.13WDD148 pKa = 4.23GNDD151 pKa = 3.24TSLLYY156 pKa = 10.46INNGLYY162 pKa = 10.57LRR164 pKa = 11.84ASAFGEE170 pKa = 4.3GGEE173 pKa = 4.34GSSAGVVEE181 pKa = 4.39MTGNTDD187 pKa = 2.86EE188 pKa = 4.18YY189 pKa = 9.52MSRR192 pKa = 11.84IVGSVSDD199 pKa = 4.4IEE201 pKa = 4.95GDD203 pKa = 3.53TYY205 pKa = 11.19TFTMPSSGDD214 pKa = 3.13TRR216 pKa = 11.84LLDD219 pKa = 4.17FSDD222 pKa = 4.05PADD225 pKa = 3.43MNGFLLTKK233 pKa = 10.73NEE235 pKa = 4.02DD236 pKa = 3.53DD237 pKa = 4.21EE238 pKa = 5.62FNGIYY243 pKa = 10.44YY244 pKa = 10.2GVSPAQGEE252 pKa = 4.53DD253 pKa = 3.55CMQSTDD259 pKa = 3.83FNNGIFRR266 pKa = 11.84AFRR269 pKa = 11.84FQCDD273 pKa = 3.69DD274 pKa = 4.16SISDD278 pKa = 3.55DD279 pKa = 3.65GLRR282 pKa = 11.84VSAYY286 pKa = 8.27IGRR289 pKa = 11.84YY290 pKa = 5.28TVSGNQMRR298 pKa = 11.84LLYY301 pKa = 10.07TYY303 pKa = 9.98AAPDD307 pKa = 3.95DD308 pKa = 4.58DD309 pKa = 4.58LAGAEE314 pKa = 4.44VVFTYY319 pKa = 9.71TRR321 pKa = 11.84SGRR324 pKa = 11.84QLTLTGSVGGSDD336 pKa = 3.51EE337 pKa = 4.17FTITMNRR344 pKa = 11.84ADD346 pKa = 4.72RR347 pKa = 11.84APAATVVNITVSGDD361 pKa = 3.31VNTFDD366 pKa = 5.44ADD368 pKa = 3.31AFLAGLAQVLDD379 pKa = 3.7IPVNTIEE386 pKa = 4.54VVSISPGSVVVVVAIRR402 pKa = 11.84EE403 pKa = 4.15DD404 pKa = 3.81ASDD407 pKa = 3.39SSAPSLSTIIKK418 pKa = 10.03RR419 pKa = 11.84IEE421 pKa = 3.95AVPAGTSIGGYY432 pKa = 8.85GLEE435 pKa = 4.2SAQASRR441 pKa = 11.84DD442 pKa = 3.82PGSTSAASVATTSWFAAAILFMAVVSQLLAA472 pKa = 4.53
Molecular weight: 49.71 kDa
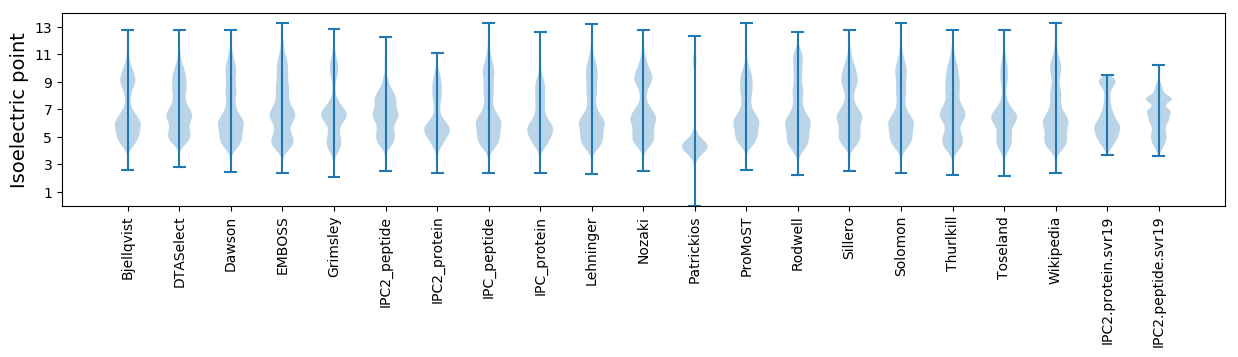
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F2UC10|F2UC10_SALR5 Gyltl1b protein OS=Salpingoeca rosetta (strain ATCC 50818 / BSB-021) OX=946362 GN=PTSG_06127 PE=4 SV=1
MM1 pKa = 6.93TMRR4 pKa = 11.84MRR6 pKa = 11.84RR7 pKa = 11.84MTFGTLVCAVAMALALASAGCAAGKK32 pKa = 10.02NGRR35 pKa = 11.84KK36 pKa = 9.17RR37 pKa = 11.84RR38 pKa = 11.84LLAFFGLVACKK49 pKa = 9.22QRR51 pKa = 11.84KK52 pKa = 6.33QRR54 pKa = 11.84KK55 pKa = 5.94QRR57 pKa = 11.84KK58 pKa = 5.93QRR60 pKa = 11.84KK61 pKa = 5.93QRR63 pKa = 11.84KK64 pKa = 5.93QRR66 pKa = 11.84KK67 pKa = 5.93QRR69 pKa = 11.84KK70 pKa = 5.93QRR72 pKa = 11.84KK73 pKa = 5.93QRR75 pKa = 11.84KK76 pKa = 5.93QRR78 pKa = 11.84KK79 pKa = 5.93QRR81 pKa = 11.84KK82 pKa = 5.93QRR84 pKa = 11.84KK85 pKa = 5.93QRR87 pKa = 11.84KK88 pKa = 5.93QRR90 pKa = 11.84KK91 pKa = 5.93QRR93 pKa = 11.84KK94 pKa = 5.93QRR96 pKa = 11.84KK97 pKa = 5.93QRR99 pKa = 11.84KK100 pKa = 5.93QRR102 pKa = 11.84KK103 pKa = 5.93QRR105 pKa = 11.84KK106 pKa = 5.93QRR108 pKa = 11.84KK109 pKa = 5.93QRR111 pKa = 11.84KK112 pKa = 5.93QRR114 pKa = 11.84KK115 pKa = 5.93QRR117 pKa = 11.84KK118 pKa = 5.93QRR120 pKa = 11.84KK121 pKa = 5.93QRR123 pKa = 11.84KK124 pKa = 5.93QRR126 pKa = 11.84KK127 pKa = 5.93QRR129 pKa = 11.84KK130 pKa = 5.93QRR132 pKa = 11.84KK133 pKa = 5.93QRR135 pKa = 11.84KK136 pKa = 6.88QRR138 pKa = 11.84KK139 pKa = 5.96QCKK142 pKa = 6.87QHH144 pKa = 5.83
MM1 pKa = 6.93TMRR4 pKa = 11.84MRR6 pKa = 11.84RR7 pKa = 11.84MTFGTLVCAVAMALALASAGCAAGKK32 pKa = 10.02NGRR35 pKa = 11.84KK36 pKa = 9.17RR37 pKa = 11.84RR38 pKa = 11.84LLAFFGLVACKK49 pKa = 9.22QRR51 pKa = 11.84KK52 pKa = 6.33QRR54 pKa = 11.84KK55 pKa = 5.94QRR57 pKa = 11.84KK58 pKa = 5.93QRR60 pKa = 11.84KK61 pKa = 5.93QRR63 pKa = 11.84KK64 pKa = 5.93QRR66 pKa = 11.84KK67 pKa = 5.93QRR69 pKa = 11.84KK70 pKa = 5.93QRR72 pKa = 11.84KK73 pKa = 5.93QRR75 pKa = 11.84KK76 pKa = 5.93QRR78 pKa = 11.84KK79 pKa = 5.93QRR81 pKa = 11.84KK82 pKa = 5.93QRR84 pKa = 11.84KK85 pKa = 5.93QRR87 pKa = 11.84KK88 pKa = 5.93QRR90 pKa = 11.84KK91 pKa = 5.93QRR93 pKa = 11.84KK94 pKa = 5.93QRR96 pKa = 11.84KK97 pKa = 5.93QRR99 pKa = 11.84KK100 pKa = 5.93QRR102 pKa = 11.84KK103 pKa = 5.93QRR105 pKa = 11.84KK106 pKa = 5.93QRR108 pKa = 11.84KK109 pKa = 5.93QRR111 pKa = 11.84KK112 pKa = 5.93QRR114 pKa = 11.84KK115 pKa = 5.93QRR117 pKa = 11.84KK118 pKa = 5.93QRR120 pKa = 11.84KK121 pKa = 5.93QRR123 pKa = 11.84KK124 pKa = 5.93QRR126 pKa = 11.84KK127 pKa = 5.93QRR129 pKa = 11.84KK130 pKa = 5.93QRR132 pKa = 11.84KK133 pKa = 5.93QRR135 pKa = 11.84KK136 pKa = 6.88QRR138 pKa = 11.84KK139 pKa = 5.96QCKK142 pKa = 6.87QHH144 pKa = 5.83
Molecular weight: 18.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
7789328 |
50 |
12309 |
665.9 |
72.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.363 ± 0.028 | 1.727 ± 0.02 |
6.517 ± 0.021 | 5.612 ± 0.03 |
3.082 ± 0.012 | 6.796 ± 0.026 |
3.185 ± 0.018 | 3.279 ± 0.015 |
3.879 ± 0.028 | 8.333 ± 0.028 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.093 ± 0.011 | 3.158 ± 0.012 |
5.266 ± 0.022 | 5.444 ± 0.033 |
6.484 ± 0.024 | 7.917 ± 0.025 |
7.069 ± 0.032 | 6.763 ± 0.031 |
1.012 ± 0.007 | 2.019 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
