
Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Sordariomycetidae; Sordariales; Sordariaceae; Neurospora; Neurospora crassa
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
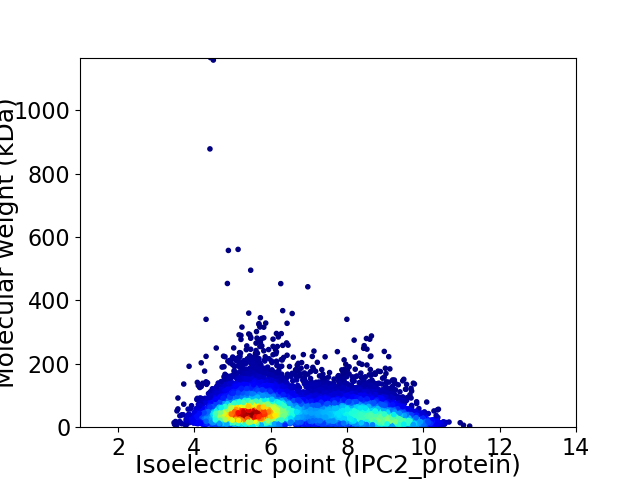
Virtual 2D-PAGE plot for 10267 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
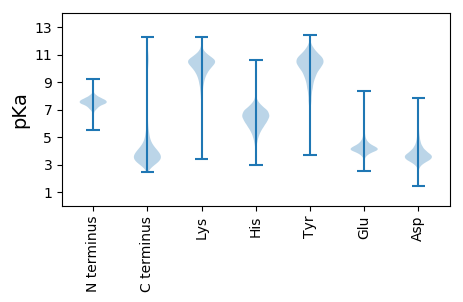
Protein with the lowest isoelectric point:
>tr|A7UW65|A7UW65_NEUCR MFS transporter Fmp42 OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=NCU10987 PE=4 SV=2
MM1 pKa = 7.53GWQSSPGRR9 pKa = 11.84LVVAAAGLFGFVNGQAYY26 pKa = 9.94FGTYY30 pKa = 9.23QDD32 pKa = 4.83TITKK36 pKa = 9.99CGSDD40 pKa = 2.91NFIYY44 pKa = 10.34LGCYY48 pKa = 10.55GNFLANAGDD57 pKa = 4.17FFRR60 pKa = 11.84FGPEE64 pKa = 3.79GYY66 pKa = 9.91NAADD70 pKa = 4.12LSLSFPGWDD79 pKa = 4.34PGSDD83 pKa = 3.05WDD85 pKa = 4.33STQTPLDD92 pKa = 3.54CARR95 pKa = 11.84TCRR98 pKa = 11.84AYY100 pKa = 10.25GYY102 pKa = 9.51KK103 pKa = 9.96YY104 pKa = 10.93SAMRR108 pKa = 11.84DD109 pKa = 3.67NNCNCGLQLPAAAVPNSDD127 pKa = 4.48SSACNAQCSGDD138 pKa = 3.69STQTCGGGLNAQVYY152 pKa = 10.02LDD154 pKa = 4.34PSFAAPDD161 pKa = 3.57QVPITPRR168 pKa = 11.84NADD171 pKa = 3.25LGSTYY176 pKa = 10.85KK177 pKa = 10.76HH178 pKa = 6.41LGCYY182 pKa = 9.04SSRR185 pKa = 11.84GFATQDD191 pKa = 3.25DD192 pKa = 4.27VRR194 pKa = 11.84ARR196 pKa = 11.84PLVDD200 pKa = 5.93DD201 pKa = 4.79IEE203 pKa = 4.95TCFEE207 pKa = 3.96TCAGLGYY214 pKa = 9.69PLVFGARR221 pKa = 11.84EE222 pKa = 4.05GAQIRR227 pKa = 11.84CQCGTNFHH235 pKa = 6.6SGDD238 pKa = 3.43YY239 pKa = 10.14RR240 pKa = 11.84IPPSSLPNPGDD251 pKa = 3.91CNALCTADD259 pKa = 4.72DD260 pKa = 4.25TNGDD264 pKa = 4.6CDD266 pKa = 4.64PLTQKK271 pKa = 10.64CCGRR275 pKa = 11.84NEE277 pKa = 4.21HH278 pKa = 6.18YY279 pKa = 9.9PVYY282 pKa = 10.54VSSEE286 pKa = 3.93LQGCYY291 pKa = 8.49VPRR294 pKa = 11.84TPGLKK299 pKa = 9.93QSADD303 pKa = 3.39QAGFACYY310 pKa = 10.02NPPSSLLGPPKK321 pKa = 10.72VLTTSLSSVNQDD333 pKa = 3.13NIIASRR339 pKa = 11.84PALVRR344 pKa = 11.84PLTVTTGTQTWYY356 pKa = 11.39LNGCYY361 pKa = 10.29GNVPAQVLDD370 pKa = 4.22LSAPSGQIISDD381 pKa = 4.38LATGTLEE388 pKa = 3.77EE389 pKa = 4.35CARR392 pKa = 11.84RR393 pKa = 11.84CSSDD397 pKa = 2.92TTFNYY402 pKa = 10.15FGMVNGRR409 pKa = 11.84DD410 pKa = 4.17CYY412 pKa = 10.98CSNTLLTPGQVDD424 pKa = 3.49DD425 pKa = 4.6MGTCRR430 pKa = 11.84VPCQDD435 pKa = 4.47DD436 pKa = 3.53ATRR439 pKa = 11.84SCGGATTPVVYY450 pKa = 10.42SLSQSGVYY458 pKa = 7.61TEE460 pKa = 4.35RR461 pKa = 11.84LANIGAGFPTYY472 pKa = 9.99TCTPSRR478 pKa = 11.84RR479 pKa = 11.84VSNDD483 pKa = 2.14AAAPITCPDD492 pKa = 4.12DD493 pKa = 3.77NGISITTAQDD503 pKa = 2.58KK504 pKa = 9.91MYY506 pKa = 10.59HH507 pKa = 5.4VDD509 pKa = 4.87CGIEE513 pKa = 4.14YY514 pKa = 9.95PDD516 pKa = 4.48GDD518 pKa = 4.59LGTVIVDD525 pKa = 4.0SYY527 pKa = 11.53AEE529 pKa = 4.08CAAACDD535 pKa = 4.26ANIDD539 pKa = 4.05CVGFIYY545 pKa = 10.17QHH547 pKa = 6.48GPANSNGLDD556 pKa = 3.5VPCYY560 pKa = 9.99LKK562 pKa = 10.94RR563 pKa = 11.84EE564 pKa = 4.32GAVTAAVSGDD574 pKa = 3.13TGNWGGRR581 pKa = 11.84FTGFADD587 pKa = 4.56GASSGSDD594 pKa = 2.79SGTGSGSDD602 pKa = 3.55SGSGTDD608 pKa = 3.86PDD610 pKa = 4.18SGSGSGTGTDD620 pKa = 3.88PDD622 pKa = 4.08SGSGSGTGTDD632 pKa = 3.88PDD634 pKa = 4.08SGSGSGTGTDD644 pKa = 3.88PDD646 pKa = 4.08SGSGSGTGTDD656 pKa = 3.88PDD658 pKa = 4.19SGSGTGTGTDD668 pKa = 3.73PDD670 pKa = 4.19SGSGSGTGTDD680 pKa = 3.88PDD682 pKa = 4.19SGSGTGTGTDD692 pKa = 3.73PDD694 pKa = 4.16SGSGSGSGTDD704 pKa = 3.64PDD706 pKa = 4.23SGSGSGDD713 pKa = 3.04GSDD716 pKa = 4.05PGTGSGSGSGNGSGSGTGNGSVVNAIFDD744 pKa = 4.09KK745 pKa = 11.21VFGPAFQGVFSSFGKK760 pKa = 10.46IFDD763 pKa = 4.03NNPFFKK769 pKa = 10.0PKK771 pKa = 10.09KK772 pKa = 8.17PAGSTRR778 pKa = 11.84AVSTGNDD785 pKa = 3.29DD786 pKa = 3.98VQDD789 pKa = 4.51LEE791 pKa = 4.72TLASDD796 pKa = 4.43PSDD799 pKa = 3.57STATRR804 pKa = 11.84SDD806 pKa = 3.26AFIGDD811 pKa = 3.66VLSNLNLGSLMSAVPGLEE829 pKa = 4.01DD830 pKa = 3.49VLGGQDD836 pKa = 4.37LGSLIPGNIRR846 pKa = 11.84APDD849 pKa = 4.0PATPPMSANLDD860 pKa = 3.73GLASLASTVLAALNTLPSDD879 pKa = 3.55IAAPGPVDD887 pKa = 3.66TDD889 pKa = 3.72SLITLASAITDD900 pKa = 3.98ALTTAADD907 pKa = 3.98GSGPGVINGKK917 pKa = 9.9DD918 pKa = 3.21FAGILNSVFDD928 pKa = 4.77SINGIGSIFPKK939 pKa = 10.44KK940 pKa = 9.12STSSSAQPSMTVAPTQSLRR959 pKa = 11.84ARR961 pKa = 11.84DD962 pKa = 3.62VAHH965 pKa = 7.28IGPHH969 pKa = 5.17PTGVLPPGNGPAEE982 pKa = 4.28VCDD985 pKa = 3.64IATLALPANIFAPTPPPDD1003 pKa = 3.9AGRR1006 pKa = 11.84CIPIIGYY1013 pKa = 8.81WYY1015 pKa = 10.55VGEE1018 pKa = 4.35TTLLPCSEE1026 pKa = 4.89GYY1028 pKa = 10.26GAGGPQEE1035 pKa = 4.42TGSPQAPGFSGGPGGHH1051 pKa = 7.3PPPPPPAGPPPAVVIPVAGSIVLTQEE1077 pKa = 4.09QLDD1080 pKa = 3.94HH1081 pKa = 7.38LLEE1084 pKa = 4.32STNIEE1089 pKa = 4.25LEE1091 pKa = 4.13WTSEE1095 pKa = 3.99GEE1097 pKa = 4.05RR1098 pKa = 11.84VLNIPGILDD1107 pKa = 3.47MSDD1110 pKa = 2.91SHH1112 pKa = 8.75SDD1114 pKa = 3.41TFRR1117 pKa = 11.84LLLDD1121 pKa = 3.48QSSDD1125 pKa = 3.76SLSHH1129 pKa = 5.76STNQFEE1135 pKa = 4.41IPISGEE1141 pKa = 3.8VGLTQSQLDD1150 pKa = 3.94SLLQDD1155 pKa = 3.26STLDD1159 pKa = 3.6IFWNSQGAKK1168 pKa = 8.95EE1169 pKa = 3.93LNIPGVINYY1178 pKa = 9.53SSSRR1182 pKa = 11.84NDD1184 pKa = 3.27TFSLRR1189 pKa = 11.84ISSSNHH1195 pKa = 4.95HH1196 pKa = 5.55TTSTASIPGPTEE1208 pKa = 3.71SSTPPAAGGEE1218 pKa = 4.41TPDD1221 pKa = 4.66GSGADD1226 pKa = 3.66EE1227 pKa = 5.0DD1228 pKa = 4.36GGYY1231 pKa = 10.93DD1232 pKa = 4.09DD1233 pKa = 6.27VEE1235 pKa = 5.98AEE1237 pKa = 4.26LAALDD1242 pKa = 4.06ALIASFGDD1250 pKa = 2.93IDD1252 pKa = 5.12FDD1254 pKa = 5.27VGDD1257 pKa = 4.94LDD1259 pKa = 4.65IPDD1262 pKa = 4.04IDD1264 pKa = 4.6VEE1266 pKa = 4.36GLDD1269 pKa = 4.13FPTVPSIDD1277 pKa = 3.79TDD1279 pKa = 3.96DD1280 pKa = 5.53LDD1282 pKa = 4.69VDD1284 pKa = 3.74VDD1286 pKa = 4.02DD1287 pKa = 4.94VDD1289 pKa = 3.8VSRR1292 pKa = 11.84VEE1294 pKa = 4.51APEE1297 pKa = 4.17SPEE1300 pKa = 4.03VPDD1303 pKa = 3.57VDD1305 pKa = 5.97AIPAPTIDD1313 pKa = 3.13VDD1315 pKa = 4.38GEE1317 pKa = 4.57VQVGLDD1323 pKa = 4.14DD1324 pKa = 4.12EE1325 pKa = 5.38SGVPVVDD1332 pKa = 4.39PVDD1335 pKa = 3.44EE1336 pKa = 4.28ATT1338 pKa = 4.18
MM1 pKa = 7.53GWQSSPGRR9 pKa = 11.84LVVAAAGLFGFVNGQAYY26 pKa = 9.94FGTYY30 pKa = 9.23QDD32 pKa = 4.83TITKK36 pKa = 9.99CGSDD40 pKa = 2.91NFIYY44 pKa = 10.34LGCYY48 pKa = 10.55GNFLANAGDD57 pKa = 4.17FFRR60 pKa = 11.84FGPEE64 pKa = 3.79GYY66 pKa = 9.91NAADD70 pKa = 4.12LSLSFPGWDD79 pKa = 4.34PGSDD83 pKa = 3.05WDD85 pKa = 4.33STQTPLDD92 pKa = 3.54CARR95 pKa = 11.84TCRR98 pKa = 11.84AYY100 pKa = 10.25GYY102 pKa = 9.51KK103 pKa = 9.96YY104 pKa = 10.93SAMRR108 pKa = 11.84DD109 pKa = 3.67NNCNCGLQLPAAAVPNSDD127 pKa = 4.48SSACNAQCSGDD138 pKa = 3.69STQTCGGGLNAQVYY152 pKa = 10.02LDD154 pKa = 4.34PSFAAPDD161 pKa = 3.57QVPITPRR168 pKa = 11.84NADD171 pKa = 3.25LGSTYY176 pKa = 10.85KK177 pKa = 10.76HH178 pKa = 6.41LGCYY182 pKa = 9.04SSRR185 pKa = 11.84GFATQDD191 pKa = 3.25DD192 pKa = 4.27VRR194 pKa = 11.84ARR196 pKa = 11.84PLVDD200 pKa = 5.93DD201 pKa = 4.79IEE203 pKa = 4.95TCFEE207 pKa = 3.96TCAGLGYY214 pKa = 9.69PLVFGARR221 pKa = 11.84EE222 pKa = 4.05GAQIRR227 pKa = 11.84CQCGTNFHH235 pKa = 6.6SGDD238 pKa = 3.43YY239 pKa = 10.14RR240 pKa = 11.84IPPSSLPNPGDD251 pKa = 3.91CNALCTADD259 pKa = 4.72DD260 pKa = 4.25TNGDD264 pKa = 4.6CDD266 pKa = 4.64PLTQKK271 pKa = 10.64CCGRR275 pKa = 11.84NEE277 pKa = 4.21HH278 pKa = 6.18YY279 pKa = 9.9PVYY282 pKa = 10.54VSSEE286 pKa = 3.93LQGCYY291 pKa = 8.49VPRR294 pKa = 11.84TPGLKK299 pKa = 9.93QSADD303 pKa = 3.39QAGFACYY310 pKa = 10.02NPPSSLLGPPKK321 pKa = 10.72VLTTSLSSVNQDD333 pKa = 3.13NIIASRR339 pKa = 11.84PALVRR344 pKa = 11.84PLTVTTGTQTWYY356 pKa = 11.39LNGCYY361 pKa = 10.29GNVPAQVLDD370 pKa = 4.22LSAPSGQIISDD381 pKa = 4.38LATGTLEE388 pKa = 3.77EE389 pKa = 4.35CARR392 pKa = 11.84RR393 pKa = 11.84CSSDD397 pKa = 2.92TTFNYY402 pKa = 10.15FGMVNGRR409 pKa = 11.84DD410 pKa = 4.17CYY412 pKa = 10.98CSNTLLTPGQVDD424 pKa = 3.49DD425 pKa = 4.6MGTCRR430 pKa = 11.84VPCQDD435 pKa = 4.47DD436 pKa = 3.53ATRR439 pKa = 11.84SCGGATTPVVYY450 pKa = 10.42SLSQSGVYY458 pKa = 7.61TEE460 pKa = 4.35RR461 pKa = 11.84LANIGAGFPTYY472 pKa = 9.99TCTPSRR478 pKa = 11.84RR479 pKa = 11.84VSNDD483 pKa = 2.14AAAPITCPDD492 pKa = 4.12DD493 pKa = 3.77NGISITTAQDD503 pKa = 2.58KK504 pKa = 9.91MYY506 pKa = 10.59HH507 pKa = 5.4VDD509 pKa = 4.87CGIEE513 pKa = 4.14YY514 pKa = 9.95PDD516 pKa = 4.48GDD518 pKa = 4.59LGTVIVDD525 pKa = 4.0SYY527 pKa = 11.53AEE529 pKa = 4.08CAAACDD535 pKa = 4.26ANIDD539 pKa = 4.05CVGFIYY545 pKa = 10.17QHH547 pKa = 6.48GPANSNGLDD556 pKa = 3.5VPCYY560 pKa = 9.99LKK562 pKa = 10.94RR563 pKa = 11.84EE564 pKa = 4.32GAVTAAVSGDD574 pKa = 3.13TGNWGGRR581 pKa = 11.84FTGFADD587 pKa = 4.56GASSGSDD594 pKa = 2.79SGTGSGSDD602 pKa = 3.55SGSGTDD608 pKa = 3.86PDD610 pKa = 4.18SGSGSGTGTDD620 pKa = 3.88PDD622 pKa = 4.08SGSGSGTGTDD632 pKa = 3.88PDD634 pKa = 4.08SGSGSGTGTDD644 pKa = 3.88PDD646 pKa = 4.08SGSGSGTGTDD656 pKa = 3.88PDD658 pKa = 4.19SGSGTGTGTDD668 pKa = 3.73PDD670 pKa = 4.19SGSGSGTGTDD680 pKa = 3.88PDD682 pKa = 4.19SGSGTGTGTDD692 pKa = 3.73PDD694 pKa = 4.16SGSGSGSGTDD704 pKa = 3.64PDD706 pKa = 4.23SGSGSGDD713 pKa = 3.04GSDD716 pKa = 4.05PGTGSGSGSGNGSGSGTGNGSVVNAIFDD744 pKa = 4.09KK745 pKa = 11.21VFGPAFQGVFSSFGKK760 pKa = 10.46IFDD763 pKa = 4.03NNPFFKK769 pKa = 10.0PKK771 pKa = 10.09KK772 pKa = 8.17PAGSTRR778 pKa = 11.84AVSTGNDD785 pKa = 3.29DD786 pKa = 3.98VQDD789 pKa = 4.51LEE791 pKa = 4.72TLASDD796 pKa = 4.43PSDD799 pKa = 3.57STATRR804 pKa = 11.84SDD806 pKa = 3.26AFIGDD811 pKa = 3.66VLSNLNLGSLMSAVPGLEE829 pKa = 4.01DD830 pKa = 3.49VLGGQDD836 pKa = 4.37LGSLIPGNIRR846 pKa = 11.84APDD849 pKa = 4.0PATPPMSANLDD860 pKa = 3.73GLASLASTVLAALNTLPSDD879 pKa = 3.55IAAPGPVDD887 pKa = 3.66TDD889 pKa = 3.72SLITLASAITDD900 pKa = 3.98ALTTAADD907 pKa = 3.98GSGPGVINGKK917 pKa = 9.9DD918 pKa = 3.21FAGILNSVFDD928 pKa = 4.77SINGIGSIFPKK939 pKa = 10.44KK940 pKa = 9.12STSSSAQPSMTVAPTQSLRR959 pKa = 11.84ARR961 pKa = 11.84DD962 pKa = 3.62VAHH965 pKa = 7.28IGPHH969 pKa = 5.17PTGVLPPGNGPAEE982 pKa = 4.28VCDD985 pKa = 3.64IATLALPANIFAPTPPPDD1003 pKa = 3.9AGRR1006 pKa = 11.84CIPIIGYY1013 pKa = 8.81WYY1015 pKa = 10.55VGEE1018 pKa = 4.35TTLLPCSEE1026 pKa = 4.89GYY1028 pKa = 10.26GAGGPQEE1035 pKa = 4.42TGSPQAPGFSGGPGGHH1051 pKa = 7.3PPPPPPAGPPPAVVIPVAGSIVLTQEE1077 pKa = 4.09QLDD1080 pKa = 3.94HH1081 pKa = 7.38LLEE1084 pKa = 4.32STNIEE1089 pKa = 4.25LEE1091 pKa = 4.13WTSEE1095 pKa = 3.99GEE1097 pKa = 4.05RR1098 pKa = 11.84VLNIPGILDD1107 pKa = 3.47MSDD1110 pKa = 2.91SHH1112 pKa = 8.75SDD1114 pKa = 3.41TFRR1117 pKa = 11.84LLLDD1121 pKa = 3.48QSSDD1125 pKa = 3.76SLSHH1129 pKa = 5.76STNQFEE1135 pKa = 4.41IPISGEE1141 pKa = 3.8VGLTQSQLDD1150 pKa = 3.94SLLQDD1155 pKa = 3.26STLDD1159 pKa = 3.6IFWNSQGAKK1168 pKa = 8.95EE1169 pKa = 3.93LNIPGVINYY1178 pKa = 9.53SSSRR1182 pKa = 11.84NDD1184 pKa = 3.27TFSLRR1189 pKa = 11.84ISSSNHH1195 pKa = 4.95HH1196 pKa = 5.55TTSTASIPGPTEE1208 pKa = 3.71SSTPPAAGGEE1218 pKa = 4.41TPDD1221 pKa = 4.66GSGADD1226 pKa = 3.66EE1227 pKa = 5.0DD1228 pKa = 4.36GGYY1231 pKa = 10.93DD1232 pKa = 4.09DD1233 pKa = 6.27VEE1235 pKa = 5.98AEE1237 pKa = 4.26LAALDD1242 pKa = 4.06ALIASFGDD1250 pKa = 2.93IDD1252 pKa = 5.12FDD1254 pKa = 5.27VGDD1257 pKa = 4.94LDD1259 pKa = 4.65IPDD1262 pKa = 4.04IDD1264 pKa = 4.6VEE1266 pKa = 4.36GLDD1269 pKa = 4.13FPTVPSIDD1277 pKa = 3.79TDD1279 pKa = 3.96DD1280 pKa = 5.53LDD1282 pKa = 4.69VDD1284 pKa = 3.74VDD1286 pKa = 4.02DD1287 pKa = 4.94VDD1289 pKa = 3.8VSRR1292 pKa = 11.84VEE1294 pKa = 4.51APEE1297 pKa = 4.17SPEE1300 pKa = 4.03VPDD1303 pKa = 3.57VDD1305 pKa = 5.97AIPAPTIDD1313 pKa = 3.13VDD1315 pKa = 4.38GEE1317 pKa = 4.57VQVGLDD1323 pKa = 4.14DD1324 pKa = 4.12EE1325 pKa = 5.38SGVPVVDD1332 pKa = 4.39PVDD1335 pKa = 3.44EE1336 pKa = 4.28ATT1338 pKa = 4.18
Molecular weight: 136.26 kDa
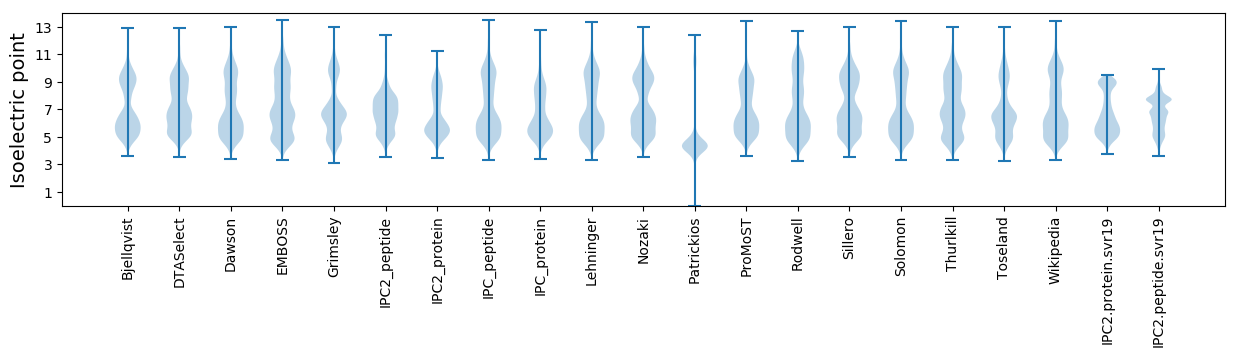
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|V5IKN2|V5IKN2_NEUCR Uncharacterized protein OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=NCU17087 PE=4 SV=1
MM1 pKa = 7.1RR2 pKa = 11.84AKK4 pKa = 9.09WRR6 pKa = 11.84KK7 pKa = 8.87KK8 pKa = 8.73RR9 pKa = 11.84VRR11 pKa = 11.84RR12 pKa = 11.84LKK14 pKa = 9.94RR15 pKa = 11.84KK16 pKa = 7.8RR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 8.6MRR21 pKa = 11.84ARR23 pKa = 11.84SKK25 pKa = 11.11
MM1 pKa = 7.1RR2 pKa = 11.84AKK4 pKa = 9.09WRR6 pKa = 11.84KK7 pKa = 8.87KK8 pKa = 8.73RR9 pKa = 11.84VRR11 pKa = 11.84RR12 pKa = 11.84LKK14 pKa = 9.94RR15 pKa = 11.84KK16 pKa = 7.8RR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 8.6MRR21 pKa = 11.84ARR23 pKa = 11.84SKK25 pKa = 11.11
Molecular weight: 3.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5305852 |
24 |
10739 |
516.8 |
56.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.676 ± 0.023 | 1.112 ± 0.009 |
5.642 ± 0.018 | 6.486 ± 0.027 |
3.389 ± 0.014 | 7.144 ± 0.024 |
2.457 ± 0.01 | 4.43 ± 0.013 |
5.076 ± 0.025 | 8.345 ± 0.028 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.151 ± 0.009 | 3.711 ± 0.016 |
6.577 ± 0.034 | 4.279 ± 0.023 |
6.148 ± 0.02 | 8.36 ± 0.031 |
6.138 ± 0.017 | 5.969 ± 0.018 |
1.344 ± 0.009 | 2.564 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
