
Panax notoginseng virus B
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; unclassified Totiviridae; Panax notoginseng virus A
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z4PGW3|A0A2Z4PGW3_9VIRU RNA-directed RNA polymerase OS=Panax notoginseng virus B OX=2248770 PE=3 SV=1
MM1 pKa = 8.06DD2 pKa = 4.56SFVKK6 pKa = 10.55QFFTEE11 pKa = 4.5FDD13 pKa = 3.7SPGVDD18 pKa = 4.25FNFDD22 pKa = 3.6SDD24 pKa = 4.69LKK26 pKa = 7.94THH28 pKa = 6.63TYY30 pKa = 8.2MGCGLRR36 pKa = 11.84VYY38 pKa = 10.18DD39 pKa = 3.44GHH41 pKa = 7.87INQSRR46 pKa = 11.84HH47 pKa = 3.57FHH49 pKa = 6.71AFQPMTPLGYY59 pKa = 10.23YY60 pKa = 10.12DD61 pKa = 4.73CEE63 pKa = 4.36VTGGGSSLDD72 pKa = 4.19GITKK76 pKa = 10.44NYY78 pKa = 7.14LTPEE82 pKa = 3.98GTVNLVTVADD92 pKa = 4.12TLKK95 pKa = 10.8GSSGLQTAMLSAHH108 pKa = 6.03VSHH111 pKa = 6.18MQHH114 pKa = 6.46WSWADD119 pKa = 3.15NHH121 pKa = 6.69VGLLFNMFRR130 pKa = 11.84YY131 pKa = 10.05VLIEE135 pKa = 4.38KK136 pKa = 9.92IRR138 pKa = 11.84EE139 pKa = 4.15CAGSLDD145 pKa = 4.0GKK147 pKa = 10.69LGLYY151 pKa = 10.27EE152 pKa = 5.11DD153 pKa = 4.08GHH155 pKa = 8.16VGIDD159 pKa = 3.31MDD161 pKa = 4.63QFWEE165 pKa = 4.78DD166 pKa = 4.62EE167 pKa = 4.22YY168 pKa = 11.73CEE170 pKa = 5.05DD171 pKa = 3.64GGARR175 pKa = 11.84KK176 pKa = 8.74WPGGDD181 pKa = 2.9VDD183 pKa = 5.49ASYY186 pKa = 11.25PDD188 pKa = 3.91YY189 pKa = 11.36LRR191 pKa = 11.84LTNTTPSTGDD201 pKa = 2.83WALDD205 pKa = 3.43LRR207 pKa = 11.84GMNARR212 pKa = 11.84DD213 pKa = 3.46ARR215 pKa = 11.84FVLLMMGAWKK225 pKa = 10.15RR226 pKa = 11.84RR227 pKa = 11.84SRR229 pKa = 11.84LRR231 pKa = 11.84LDD233 pKa = 3.38CTTKK237 pKa = 10.77KK238 pKa = 10.8LCDD241 pKa = 3.43CVLYY245 pKa = 10.51RR246 pKa = 11.84SSVEE250 pKa = 3.95VANITDD256 pKa = 4.5WIVDD260 pKa = 3.63KK261 pKa = 10.95EE262 pKa = 4.17KK263 pKa = 11.22GIVPPTLSSSEE274 pKa = 3.47AWQALKK280 pKa = 10.5FYY282 pKa = 8.93VTQNRR287 pKa = 11.84LFDD290 pKa = 4.06HH291 pKa = 7.0FSATLYY297 pKa = 10.45IIATMMYY304 pKa = 9.52QFVPATAEE312 pKa = 3.76ATNWLRR318 pKa = 11.84LKK320 pKa = 8.83WTVNIPTFRR329 pKa = 11.84SVRR332 pKa = 11.84GRR334 pKa = 11.84YY335 pKa = 8.47EE336 pKa = 3.67FLNEE340 pKa = 4.17GEE342 pKa = 4.36PALLSHH348 pKa = 7.09RR349 pKa = 11.84CLNEE353 pKa = 3.06WGYY356 pKa = 11.01INGKK360 pKa = 6.53MEE362 pKa = 4.51KK363 pKa = 9.35VNLMALVFSQAYY375 pKa = 6.63HH376 pKa = 6.12TGLAVRR382 pKa = 11.84GVRR385 pKa = 11.84KK386 pKa = 9.83GLEE389 pKa = 3.98LHH391 pKa = 7.07PDD393 pKa = 4.1DD394 pKa = 5.02IFSSEE399 pKa = 3.31IDD401 pKa = 3.97FYY403 pKa = 11.35TSANFISAAASEE415 pKa = 4.55ATRR418 pKa = 11.84TDD420 pKa = 3.11TPLPGMVGIHH430 pKa = 5.71FVVNDD435 pKa = 3.9DD436 pKa = 3.72FDD438 pKa = 5.38VYY440 pKa = 11.41DD441 pKa = 3.71EE442 pKa = 4.0NRR444 pKa = 11.84RR445 pKa = 11.84IEE447 pKa = 4.05TRR449 pKa = 11.84EE450 pKa = 3.65NDD452 pKa = 3.95DD453 pKa = 3.38EE454 pKa = 5.66HH455 pKa = 8.68LDD457 pKa = 4.21DD458 pKa = 4.13YY459 pKa = 10.7HH460 pKa = 5.99TSKK463 pKa = 11.3DD464 pKa = 3.41KK465 pKa = 11.53VSVKK469 pKa = 10.35KK470 pKa = 10.97GSTLLVDD477 pKa = 4.54VNDD480 pKa = 4.1STLSRR485 pKa = 11.84AALDD489 pKa = 4.77LISKK493 pKa = 9.91KK494 pKa = 10.33IKK496 pKa = 9.48TLMKK500 pKa = 9.96GVRR503 pKa = 11.84AKK505 pKa = 11.13LEE507 pKa = 4.33DD508 pKa = 4.12DD509 pKa = 3.69VDD511 pKa = 4.75FEE513 pKa = 4.62LTDD516 pKa = 3.71EE517 pKa = 4.54EE518 pKa = 5.13KK519 pKa = 10.78LAQEE523 pKa = 4.03GKK525 pKa = 9.93MYY527 pKa = 9.56VTLAADD533 pKa = 3.68EE534 pKa = 4.15QEE536 pKa = 4.16QMEE539 pKa = 4.7EE540 pKa = 4.09KK541 pKa = 10.87VFVHH545 pKa = 5.78MPWYY549 pKa = 8.26TFPGVPTMILPLSPFPYY566 pKa = 8.96NTPFNMKK573 pKa = 9.91GRR575 pKa = 11.84ITTDD579 pKa = 2.15IGKK582 pKa = 9.54VDD584 pKa = 3.38RR585 pKa = 11.84RR586 pKa = 11.84GLRR589 pKa = 11.84LVSKK593 pKa = 7.45TAWEE597 pKa = 4.35VANLMRR603 pKa = 11.84LCGYY607 pKa = 10.22DD608 pKa = 3.06SRR610 pKa = 11.84FRR612 pKa = 11.84DD613 pKa = 3.52GGEE616 pKa = 3.5IAGAGTYY623 pKa = 9.04FSPNDD628 pKa = 3.76ANMVWPVLQVPDD640 pKa = 3.97AQDD643 pKa = 3.78DD644 pKa = 4.18EE645 pKa = 5.47VILTGQIDD653 pKa = 3.76RR654 pKa = 11.84EE655 pKa = 4.29AQFIDD660 pKa = 4.51LPPMFNRR667 pKa = 11.84FFIGKK672 pKa = 8.17EE673 pKa = 3.62VEE675 pKa = 4.03YY676 pKa = 10.0SIKK679 pKa = 10.37VIRR682 pKa = 11.84RR683 pKa = 11.84GTATSFVDD691 pKa = 3.2NRR693 pKa = 11.84KK694 pKa = 9.8DD695 pKa = 3.37VAEE698 pKa = 4.08WGGAVTLTRR707 pKa = 11.84GVTVSYY713 pKa = 10.52NVPEE717 pKa = 4.23NVSKK721 pKa = 10.78LRR723 pKa = 11.84AYY725 pKa = 9.88ISRR728 pKa = 11.84NEE730 pKa = 3.87TGFRR734 pKa = 11.84FVDD737 pKa = 3.36NAQAGVIPMPEE748 pKa = 4.53PDD750 pKa = 4.5LNAQGVAGVV759 pKa = 3.57
MM1 pKa = 8.06DD2 pKa = 4.56SFVKK6 pKa = 10.55QFFTEE11 pKa = 4.5FDD13 pKa = 3.7SPGVDD18 pKa = 4.25FNFDD22 pKa = 3.6SDD24 pKa = 4.69LKK26 pKa = 7.94THH28 pKa = 6.63TYY30 pKa = 8.2MGCGLRR36 pKa = 11.84VYY38 pKa = 10.18DD39 pKa = 3.44GHH41 pKa = 7.87INQSRR46 pKa = 11.84HH47 pKa = 3.57FHH49 pKa = 6.71AFQPMTPLGYY59 pKa = 10.23YY60 pKa = 10.12DD61 pKa = 4.73CEE63 pKa = 4.36VTGGGSSLDD72 pKa = 4.19GITKK76 pKa = 10.44NYY78 pKa = 7.14LTPEE82 pKa = 3.98GTVNLVTVADD92 pKa = 4.12TLKK95 pKa = 10.8GSSGLQTAMLSAHH108 pKa = 6.03VSHH111 pKa = 6.18MQHH114 pKa = 6.46WSWADD119 pKa = 3.15NHH121 pKa = 6.69VGLLFNMFRR130 pKa = 11.84YY131 pKa = 10.05VLIEE135 pKa = 4.38KK136 pKa = 9.92IRR138 pKa = 11.84EE139 pKa = 4.15CAGSLDD145 pKa = 4.0GKK147 pKa = 10.69LGLYY151 pKa = 10.27EE152 pKa = 5.11DD153 pKa = 4.08GHH155 pKa = 8.16VGIDD159 pKa = 3.31MDD161 pKa = 4.63QFWEE165 pKa = 4.78DD166 pKa = 4.62EE167 pKa = 4.22YY168 pKa = 11.73CEE170 pKa = 5.05DD171 pKa = 3.64GGARR175 pKa = 11.84KK176 pKa = 8.74WPGGDD181 pKa = 2.9VDD183 pKa = 5.49ASYY186 pKa = 11.25PDD188 pKa = 3.91YY189 pKa = 11.36LRR191 pKa = 11.84LTNTTPSTGDD201 pKa = 2.83WALDD205 pKa = 3.43LRR207 pKa = 11.84GMNARR212 pKa = 11.84DD213 pKa = 3.46ARR215 pKa = 11.84FVLLMMGAWKK225 pKa = 10.15RR226 pKa = 11.84RR227 pKa = 11.84SRR229 pKa = 11.84LRR231 pKa = 11.84LDD233 pKa = 3.38CTTKK237 pKa = 10.77KK238 pKa = 10.8LCDD241 pKa = 3.43CVLYY245 pKa = 10.51RR246 pKa = 11.84SSVEE250 pKa = 3.95VANITDD256 pKa = 4.5WIVDD260 pKa = 3.63KK261 pKa = 10.95EE262 pKa = 4.17KK263 pKa = 11.22GIVPPTLSSSEE274 pKa = 3.47AWQALKK280 pKa = 10.5FYY282 pKa = 8.93VTQNRR287 pKa = 11.84LFDD290 pKa = 4.06HH291 pKa = 7.0FSATLYY297 pKa = 10.45IIATMMYY304 pKa = 9.52QFVPATAEE312 pKa = 3.76ATNWLRR318 pKa = 11.84LKK320 pKa = 8.83WTVNIPTFRR329 pKa = 11.84SVRR332 pKa = 11.84GRR334 pKa = 11.84YY335 pKa = 8.47EE336 pKa = 3.67FLNEE340 pKa = 4.17GEE342 pKa = 4.36PALLSHH348 pKa = 7.09RR349 pKa = 11.84CLNEE353 pKa = 3.06WGYY356 pKa = 11.01INGKK360 pKa = 6.53MEE362 pKa = 4.51KK363 pKa = 9.35VNLMALVFSQAYY375 pKa = 6.63HH376 pKa = 6.12TGLAVRR382 pKa = 11.84GVRR385 pKa = 11.84KK386 pKa = 9.83GLEE389 pKa = 3.98LHH391 pKa = 7.07PDD393 pKa = 4.1DD394 pKa = 5.02IFSSEE399 pKa = 3.31IDD401 pKa = 3.97FYY403 pKa = 11.35TSANFISAAASEE415 pKa = 4.55ATRR418 pKa = 11.84TDD420 pKa = 3.11TPLPGMVGIHH430 pKa = 5.71FVVNDD435 pKa = 3.9DD436 pKa = 3.72FDD438 pKa = 5.38VYY440 pKa = 11.41DD441 pKa = 3.71EE442 pKa = 4.0NRR444 pKa = 11.84RR445 pKa = 11.84IEE447 pKa = 4.05TRR449 pKa = 11.84EE450 pKa = 3.65NDD452 pKa = 3.95DD453 pKa = 3.38EE454 pKa = 5.66HH455 pKa = 8.68LDD457 pKa = 4.21DD458 pKa = 4.13YY459 pKa = 10.7HH460 pKa = 5.99TSKK463 pKa = 11.3DD464 pKa = 3.41KK465 pKa = 11.53VSVKK469 pKa = 10.35KK470 pKa = 10.97GSTLLVDD477 pKa = 4.54VNDD480 pKa = 4.1STLSRR485 pKa = 11.84AALDD489 pKa = 4.77LISKK493 pKa = 9.91KK494 pKa = 10.33IKK496 pKa = 9.48TLMKK500 pKa = 9.96GVRR503 pKa = 11.84AKK505 pKa = 11.13LEE507 pKa = 4.33DD508 pKa = 4.12DD509 pKa = 3.69VDD511 pKa = 4.75FEE513 pKa = 4.62LTDD516 pKa = 3.71EE517 pKa = 4.54EE518 pKa = 5.13KK519 pKa = 10.78LAQEE523 pKa = 4.03GKK525 pKa = 9.93MYY527 pKa = 9.56VTLAADD533 pKa = 3.68EE534 pKa = 4.15QEE536 pKa = 4.16QMEE539 pKa = 4.7EE540 pKa = 4.09KK541 pKa = 10.87VFVHH545 pKa = 5.78MPWYY549 pKa = 8.26TFPGVPTMILPLSPFPYY566 pKa = 8.96NTPFNMKK573 pKa = 9.91GRR575 pKa = 11.84ITTDD579 pKa = 2.15IGKK582 pKa = 9.54VDD584 pKa = 3.38RR585 pKa = 11.84RR586 pKa = 11.84GLRR589 pKa = 11.84LVSKK593 pKa = 7.45TAWEE597 pKa = 4.35VANLMRR603 pKa = 11.84LCGYY607 pKa = 10.22DD608 pKa = 3.06SRR610 pKa = 11.84FRR612 pKa = 11.84DD613 pKa = 3.52GGEE616 pKa = 3.5IAGAGTYY623 pKa = 9.04FSPNDD628 pKa = 3.76ANMVWPVLQVPDD640 pKa = 3.97AQDD643 pKa = 3.78DD644 pKa = 4.18EE645 pKa = 5.47VILTGQIDD653 pKa = 3.76RR654 pKa = 11.84EE655 pKa = 4.29AQFIDD660 pKa = 4.51LPPMFNRR667 pKa = 11.84FFIGKK672 pKa = 8.17EE673 pKa = 3.62VEE675 pKa = 4.03YY676 pKa = 10.0SIKK679 pKa = 10.37VIRR682 pKa = 11.84RR683 pKa = 11.84GTATSFVDD691 pKa = 3.2NRR693 pKa = 11.84KK694 pKa = 9.8DD695 pKa = 3.37VAEE698 pKa = 4.08WGGAVTLTRR707 pKa = 11.84GVTVSYY713 pKa = 10.52NVPEE717 pKa = 4.23NVSKK721 pKa = 10.78LRR723 pKa = 11.84AYY725 pKa = 9.88ISRR728 pKa = 11.84NEE730 pKa = 3.87TGFRR734 pKa = 11.84FVDD737 pKa = 3.36NAQAGVIPMPEE748 pKa = 4.53PDD750 pKa = 4.5LNAQGVAGVV759 pKa = 3.57
Molecular weight: 85.62 kDa
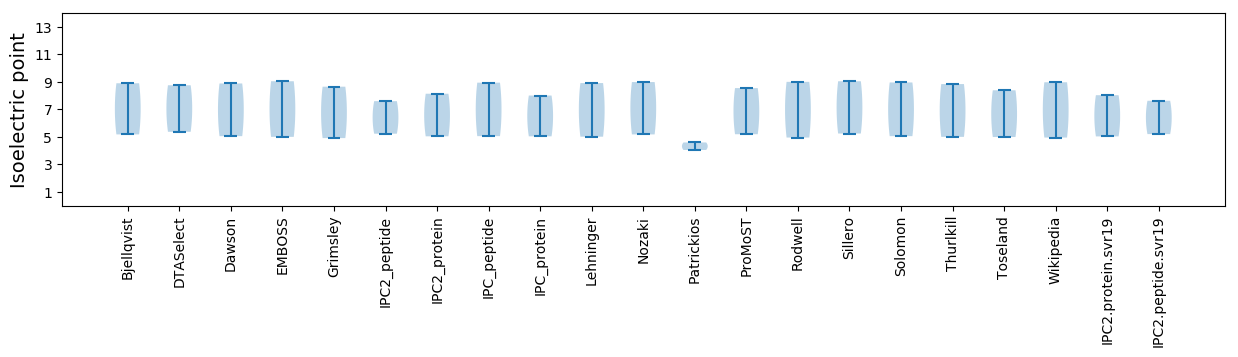
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z4PGW3|A0A2Z4PGW3_9VIRU RNA-directed RNA polymerase OS=Panax notoginseng virus B OX=2248770 PE=3 SV=1
MM1 pKa = 7.09VRR3 pKa = 11.84RR4 pKa = 11.84AMSVAYY10 pKa = 10.17SQVDD14 pKa = 3.47QYY16 pKa = 12.0DD17 pKa = 3.58YY18 pKa = 11.79SDD20 pKa = 3.13WQLFRR25 pKa = 11.84FLRR28 pKa = 11.84RR29 pKa = 11.84VFDD32 pKa = 3.56VDD34 pKa = 3.46RR35 pKa = 11.84KK36 pKa = 10.5LITHH40 pKa = 7.2ARR42 pKa = 11.84TGSGPVEE49 pKa = 4.22GEE51 pKa = 4.01FPRR54 pKa = 11.84AAITGEE60 pKa = 3.91HH61 pKa = 5.97HH62 pKa = 4.7THH64 pKa = 6.33FRR66 pKa = 11.84PEE68 pKa = 4.23EE69 pKa = 3.77VWEE72 pKa = 3.96VAKK75 pKa = 10.32IYY77 pKa = 9.79KK78 pKa = 10.28AKK80 pKa = 10.0LRR82 pKa = 11.84AMSVVYY88 pKa = 10.62RR89 pKa = 11.84NLKK92 pKa = 9.55KK93 pKa = 10.31IEE95 pKa = 4.93GITEE99 pKa = 4.16AVVSTMFLYY108 pKa = 11.02VLFARR113 pKa = 11.84PQIAYY118 pKa = 10.1LFACSRR124 pKa = 11.84RR125 pKa = 11.84IWASKK130 pKa = 10.84DD131 pKa = 3.11VGQLANVLKK140 pKa = 10.58EE141 pKa = 3.37ISTPLKK147 pKa = 10.35SVQNYY152 pKa = 7.34EE153 pKa = 4.21LSDD156 pKa = 3.47VTQLFEE162 pKa = 4.52LQCLVNRR169 pKa = 11.84GIGEE173 pKa = 4.0VDD175 pKa = 2.88WNKK178 pKa = 9.66EE179 pKa = 3.5RR180 pKa = 11.84RR181 pKa = 11.84NRR183 pKa = 11.84EE184 pKa = 3.72CPNVVKK190 pKa = 10.86VDD192 pKa = 3.54VVDD195 pKa = 4.35VYY197 pKa = 11.48KK198 pKa = 10.73EE199 pKa = 3.93AVDD202 pKa = 3.56MFRR205 pKa = 11.84MGVSHH210 pKa = 6.94GYY212 pKa = 10.06KK213 pKa = 9.16YY214 pKa = 11.47ARR216 pKa = 11.84MDD218 pKa = 3.35LKK220 pKa = 10.98KK221 pKa = 10.36YY222 pKa = 10.23IKK224 pKa = 10.33SRR226 pKa = 11.84WEE228 pKa = 3.78WVPSGSVHH236 pKa = 5.32SQYY239 pKa = 10.6IEE241 pKa = 3.94DD242 pKa = 3.44QHH244 pKa = 7.17FIKK247 pKa = 9.43TDD249 pKa = 2.78HH250 pKa = 4.95MHH252 pKa = 5.59RR253 pKa = 11.84TKK255 pKa = 10.62FVTLNMMKK263 pKa = 9.56TEE265 pKa = 3.89HH266 pKa = 6.36LRR268 pKa = 11.84RR269 pKa = 11.84MFQRR273 pKa = 11.84RR274 pKa = 11.84KK275 pKa = 9.8EE276 pKa = 3.99IQAWASVKK284 pKa = 10.46YY285 pKa = 9.66EE286 pKa = 3.86WAKK289 pKa = 10.25QRR291 pKa = 11.84AIYY294 pKa = 10.09GVDD297 pKa = 3.36LTSSVITNFAMFRR310 pKa = 11.84CEE312 pKa = 3.74EE313 pKa = 4.21VFKK316 pKa = 11.14HH317 pKa = 5.86RR318 pKa = 11.84FPVGEE323 pKa = 3.82EE324 pKa = 3.53AAASRR329 pKa = 11.84VHH331 pKa = 6.42KK332 pKa = 9.57RR333 pKa = 11.84LKK335 pKa = 10.36MMLDD339 pKa = 3.72GNEE342 pKa = 4.16SFCYY346 pKa = 10.74DD347 pKa = 3.41FDD349 pKa = 5.14DD350 pKa = 5.86FNAQHH355 pKa = 5.87STEE358 pKa = 3.87SMYY361 pKa = 11.38AVLQAYY367 pKa = 10.6LDD369 pKa = 3.83VFKK372 pKa = 11.5GNMSDD377 pKa = 4.05DD378 pKa = 3.6QVEE381 pKa = 4.01AMKK384 pKa = 10.06WVRR387 pKa = 11.84DD388 pKa = 3.8SILSVKK394 pKa = 8.96VHH396 pKa = 6.03NNEE399 pKa = 3.8KK400 pKa = 10.54NRR402 pKa = 11.84DD403 pKa = 3.37EE404 pKa = 4.32MYY406 pKa = 9.6STNGTLLSGWRR417 pKa = 11.84LTTFMNTALNYY428 pKa = 9.79IYY430 pKa = 10.46FKK432 pKa = 10.92LAGAFDD438 pKa = 3.58IAGVKK443 pKa = 10.5DD444 pKa = 3.91SVHH447 pKa = 6.57NGDD450 pKa = 4.68DD451 pKa = 3.43VLVAIKK457 pKa = 9.97DD458 pKa = 3.81LKK460 pKa = 10.03SACSIHH466 pKa = 6.44QAMAKK471 pKa = 9.75INARR475 pKa = 11.84AQATKK480 pKa = 10.86CNIFSVGEE488 pKa = 3.86FLRR491 pKa = 11.84VEE493 pKa = 4.1HH494 pKa = 6.89KK495 pKa = 10.16ISKK498 pKa = 10.16EE499 pKa = 3.82EE500 pKa = 3.96GLGAQYY506 pKa = 9.05LTRR509 pKa = 11.84AAATLAHH516 pKa = 6.4SRR518 pKa = 11.84IEE520 pKa = 4.27SQEE523 pKa = 3.57PTRR526 pKa = 11.84LVDD529 pKa = 3.96SIKK532 pKa = 11.09GMVTRR537 pKa = 11.84CEE539 pKa = 4.14EE540 pKa = 3.77ITARR544 pKa = 11.84SYY546 pKa = 12.27DD547 pKa = 3.78MVEE550 pKa = 4.0VAYY553 pKa = 10.4HH554 pKa = 7.14LLTEE558 pKa = 4.45SVTRR562 pKa = 11.84LCGIFGVKK570 pKa = 10.02KK571 pKa = 9.32EE572 pKa = 4.81DD573 pKa = 3.4GMKK576 pKa = 8.98VTRR579 pKa = 11.84SHH581 pKa = 6.77LLVGGASEE589 pKa = 4.24TFEE592 pKa = 4.94GIVDD596 pKa = 3.58EE597 pKa = 4.83TIEE600 pKa = 4.26EE601 pKa = 4.12EE602 pKa = 4.6VLIDD606 pKa = 3.38DD607 pKa = 4.5TRR609 pKa = 11.84ITEE612 pKa = 3.96EE613 pKa = 4.09NRR615 pKa = 11.84DD616 pKa = 3.18KK617 pKa = 11.07RR618 pKa = 11.84ATIRR622 pKa = 11.84EE623 pKa = 4.48LMPGINDD630 pKa = 3.3YY631 pKa = 11.75AEE633 pKa = 4.18ILEE636 pKa = 4.34TQYY639 pKa = 11.86GEE641 pKa = 4.96FMPLDD646 pKa = 4.0DD647 pKa = 4.96VRR649 pKa = 11.84KK650 pKa = 9.27KK651 pKa = 10.88VINATHH657 pKa = 6.4RR658 pKa = 11.84QLEE661 pKa = 4.32ITRR664 pKa = 11.84GTNLKK669 pKa = 8.64ITDD672 pKa = 3.23VRR674 pKa = 11.84HH675 pKa = 4.1TVKK678 pKa = 10.95YY679 pKa = 10.12KK680 pKa = 10.43FGRR683 pKa = 11.84ALFRR687 pKa = 11.84MYY689 pKa = 10.45KK690 pKa = 10.24DD691 pKa = 3.45KK692 pKa = 11.74VNIPYY697 pKa = 9.74VEE699 pKa = 4.11KK700 pKa = 10.9ARR702 pKa = 11.84FLGIPPIAMLSHH714 pKa = 7.13SSMKK718 pKa = 10.26FVTKK722 pKa = 10.64LISSVKK728 pKa = 10.58DD729 pKa = 2.94VDD731 pKa = 3.66YY732 pKa = 10.68TLRR735 pKa = 11.84ALLL738 pKa = 4.1
MM1 pKa = 7.09VRR3 pKa = 11.84RR4 pKa = 11.84AMSVAYY10 pKa = 10.17SQVDD14 pKa = 3.47QYY16 pKa = 12.0DD17 pKa = 3.58YY18 pKa = 11.79SDD20 pKa = 3.13WQLFRR25 pKa = 11.84FLRR28 pKa = 11.84RR29 pKa = 11.84VFDD32 pKa = 3.56VDD34 pKa = 3.46RR35 pKa = 11.84KK36 pKa = 10.5LITHH40 pKa = 7.2ARR42 pKa = 11.84TGSGPVEE49 pKa = 4.22GEE51 pKa = 4.01FPRR54 pKa = 11.84AAITGEE60 pKa = 3.91HH61 pKa = 5.97HH62 pKa = 4.7THH64 pKa = 6.33FRR66 pKa = 11.84PEE68 pKa = 4.23EE69 pKa = 3.77VWEE72 pKa = 3.96VAKK75 pKa = 10.32IYY77 pKa = 9.79KK78 pKa = 10.28AKK80 pKa = 10.0LRR82 pKa = 11.84AMSVVYY88 pKa = 10.62RR89 pKa = 11.84NLKK92 pKa = 9.55KK93 pKa = 10.31IEE95 pKa = 4.93GITEE99 pKa = 4.16AVVSTMFLYY108 pKa = 11.02VLFARR113 pKa = 11.84PQIAYY118 pKa = 10.1LFACSRR124 pKa = 11.84RR125 pKa = 11.84IWASKK130 pKa = 10.84DD131 pKa = 3.11VGQLANVLKK140 pKa = 10.58EE141 pKa = 3.37ISTPLKK147 pKa = 10.35SVQNYY152 pKa = 7.34EE153 pKa = 4.21LSDD156 pKa = 3.47VTQLFEE162 pKa = 4.52LQCLVNRR169 pKa = 11.84GIGEE173 pKa = 4.0VDD175 pKa = 2.88WNKK178 pKa = 9.66EE179 pKa = 3.5RR180 pKa = 11.84RR181 pKa = 11.84NRR183 pKa = 11.84EE184 pKa = 3.72CPNVVKK190 pKa = 10.86VDD192 pKa = 3.54VVDD195 pKa = 4.35VYY197 pKa = 11.48KK198 pKa = 10.73EE199 pKa = 3.93AVDD202 pKa = 3.56MFRR205 pKa = 11.84MGVSHH210 pKa = 6.94GYY212 pKa = 10.06KK213 pKa = 9.16YY214 pKa = 11.47ARR216 pKa = 11.84MDD218 pKa = 3.35LKK220 pKa = 10.98KK221 pKa = 10.36YY222 pKa = 10.23IKK224 pKa = 10.33SRR226 pKa = 11.84WEE228 pKa = 3.78WVPSGSVHH236 pKa = 5.32SQYY239 pKa = 10.6IEE241 pKa = 3.94DD242 pKa = 3.44QHH244 pKa = 7.17FIKK247 pKa = 9.43TDD249 pKa = 2.78HH250 pKa = 4.95MHH252 pKa = 5.59RR253 pKa = 11.84TKK255 pKa = 10.62FVTLNMMKK263 pKa = 9.56TEE265 pKa = 3.89HH266 pKa = 6.36LRR268 pKa = 11.84RR269 pKa = 11.84MFQRR273 pKa = 11.84RR274 pKa = 11.84KK275 pKa = 9.8EE276 pKa = 3.99IQAWASVKK284 pKa = 10.46YY285 pKa = 9.66EE286 pKa = 3.86WAKK289 pKa = 10.25QRR291 pKa = 11.84AIYY294 pKa = 10.09GVDD297 pKa = 3.36LTSSVITNFAMFRR310 pKa = 11.84CEE312 pKa = 3.74EE313 pKa = 4.21VFKK316 pKa = 11.14HH317 pKa = 5.86RR318 pKa = 11.84FPVGEE323 pKa = 3.82EE324 pKa = 3.53AAASRR329 pKa = 11.84VHH331 pKa = 6.42KK332 pKa = 9.57RR333 pKa = 11.84LKK335 pKa = 10.36MMLDD339 pKa = 3.72GNEE342 pKa = 4.16SFCYY346 pKa = 10.74DD347 pKa = 3.41FDD349 pKa = 5.14DD350 pKa = 5.86FNAQHH355 pKa = 5.87STEE358 pKa = 3.87SMYY361 pKa = 11.38AVLQAYY367 pKa = 10.6LDD369 pKa = 3.83VFKK372 pKa = 11.5GNMSDD377 pKa = 4.05DD378 pKa = 3.6QVEE381 pKa = 4.01AMKK384 pKa = 10.06WVRR387 pKa = 11.84DD388 pKa = 3.8SILSVKK394 pKa = 8.96VHH396 pKa = 6.03NNEE399 pKa = 3.8KK400 pKa = 10.54NRR402 pKa = 11.84DD403 pKa = 3.37EE404 pKa = 4.32MYY406 pKa = 9.6STNGTLLSGWRR417 pKa = 11.84LTTFMNTALNYY428 pKa = 9.79IYY430 pKa = 10.46FKK432 pKa = 10.92LAGAFDD438 pKa = 3.58IAGVKK443 pKa = 10.5DD444 pKa = 3.91SVHH447 pKa = 6.57NGDD450 pKa = 4.68DD451 pKa = 3.43VLVAIKK457 pKa = 9.97DD458 pKa = 3.81LKK460 pKa = 10.03SACSIHH466 pKa = 6.44QAMAKK471 pKa = 9.75INARR475 pKa = 11.84AQATKK480 pKa = 10.86CNIFSVGEE488 pKa = 3.86FLRR491 pKa = 11.84VEE493 pKa = 4.1HH494 pKa = 6.89KK495 pKa = 10.16ISKK498 pKa = 10.16EE499 pKa = 3.82EE500 pKa = 3.96GLGAQYY506 pKa = 9.05LTRR509 pKa = 11.84AAATLAHH516 pKa = 6.4SRR518 pKa = 11.84IEE520 pKa = 4.27SQEE523 pKa = 3.57PTRR526 pKa = 11.84LVDD529 pKa = 3.96SIKK532 pKa = 11.09GMVTRR537 pKa = 11.84CEE539 pKa = 4.14EE540 pKa = 3.77ITARR544 pKa = 11.84SYY546 pKa = 12.27DD547 pKa = 3.78MVEE550 pKa = 4.0VAYY553 pKa = 10.4HH554 pKa = 7.14LLTEE558 pKa = 4.45SVTRR562 pKa = 11.84LCGIFGVKK570 pKa = 10.02KK571 pKa = 9.32EE572 pKa = 4.81DD573 pKa = 3.4GMKK576 pKa = 8.98VTRR579 pKa = 11.84SHH581 pKa = 6.77LLVGGASEE589 pKa = 4.24TFEE592 pKa = 4.94GIVDD596 pKa = 3.58EE597 pKa = 4.83TIEE600 pKa = 4.26EE601 pKa = 4.12EE602 pKa = 4.6VLIDD606 pKa = 3.38DD607 pKa = 4.5TRR609 pKa = 11.84ITEE612 pKa = 3.96EE613 pKa = 4.09NRR615 pKa = 11.84DD616 pKa = 3.18KK617 pKa = 11.07RR618 pKa = 11.84ATIRR622 pKa = 11.84EE623 pKa = 4.48LMPGINDD630 pKa = 3.3YY631 pKa = 11.75AEE633 pKa = 4.18ILEE636 pKa = 4.34TQYY639 pKa = 11.86GEE641 pKa = 4.96FMPLDD646 pKa = 4.0DD647 pKa = 4.96VRR649 pKa = 11.84KK650 pKa = 9.27KK651 pKa = 10.88VINATHH657 pKa = 6.4RR658 pKa = 11.84QLEE661 pKa = 4.32ITRR664 pKa = 11.84GTNLKK669 pKa = 8.64ITDD672 pKa = 3.23VRR674 pKa = 11.84HH675 pKa = 4.1TVKK678 pKa = 10.95YY679 pKa = 10.12KK680 pKa = 10.43FGRR683 pKa = 11.84ALFRR687 pKa = 11.84MYY689 pKa = 10.45KK690 pKa = 10.24DD691 pKa = 3.45KK692 pKa = 11.74VNIPYY697 pKa = 9.74VEE699 pKa = 4.11KK700 pKa = 10.9ARR702 pKa = 11.84FLGIPPIAMLSHH714 pKa = 7.13SSMKK718 pKa = 10.26FVTKK722 pKa = 10.64LISSVKK728 pKa = 10.58DD729 pKa = 2.94VDD731 pKa = 3.66YY732 pKa = 10.68TLRR735 pKa = 11.84ALLL738 pKa = 4.1
Molecular weight: 85.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1497 |
738 |
759 |
748.5 |
85.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.947 ± 0.257 | 1.202 ± 0.012 |
7.014 ± 0.826 | 6.546 ± 0.536 |
4.743 ± 0.189 | 6.146 ± 0.976 |
2.672 ± 0.309 | 4.81 ± 0.519 |
5.945 ± 0.86 | 7.882 ± 0.393 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.54 ± 0.176 | 3.941 ± 0.291 |
3.006 ± 0.772 | 2.739 ± 0.169 |
6.613 ± 0.584 | 5.945 ± 0.106 |
6.279 ± 0.409 | 8.484 ± 0.414 |
1.67 ± 0.219 | 3.874 ± 0.133 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
