
Thin paspalum asymptomatic virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Procedovirinae; Panicovirus
Average proteome isoelectric point is 7.7
Get precalculated fractions of proteins
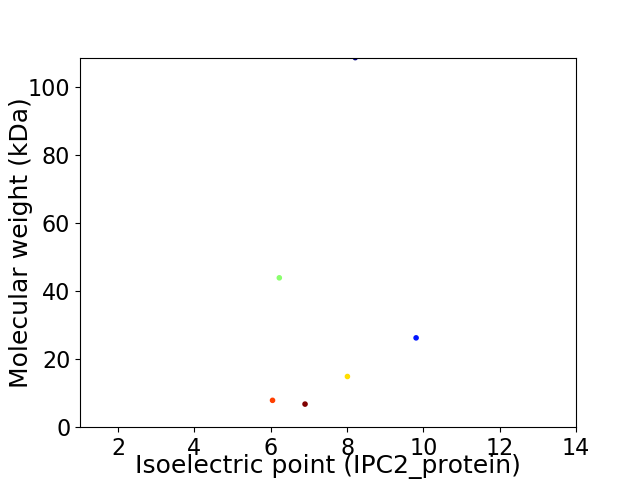
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S4WHA6|S4WHA6_9TOMB Capsid protein OS=Thin paspalum asymptomatic virus OX=1352511 PE=3 SV=1
MM1 pKa = 7.6EE2 pKa = 5.19SFVSRR7 pKa = 11.84ISEE10 pKa = 4.09ALSSCDD16 pKa = 4.77ANLDD20 pKa = 3.61YY21 pKa = 11.57DD22 pKa = 4.65KK23 pKa = 11.03MLLILKK29 pKa = 8.7HH30 pKa = 6.65CPDD33 pKa = 4.83KK34 pKa = 11.1LLSWASGIEE43 pKa = 3.95GLQTHH48 pKa = 5.38QLKK51 pKa = 10.33VAITHH56 pKa = 5.75LVKK59 pKa = 10.29TEE61 pKa = 3.53FHH63 pKa = 5.84TCKK66 pKa = 10.39HH67 pKa = 6.11LLTTMSATTADD78 pKa = 4.18YY79 pKa = 10.78IKK81 pKa = 10.35MFMEE85 pKa = 4.69LSPEE89 pKa = 3.79QEE91 pKa = 4.3VIINTGGTVSWWPKK105 pKa = 9.94LIRR108 pKa = 11.84DD109 pKa = 3.47IPEE112 pKa = 3.81WFTDD116 pKa = 4.49KK117 pKa = 10.61IPHH120 pKa = 6.57YY121 pKa = 9.81ILQIVDD127 pKa = 5.75FILRR131 pKa = 11.84WVSRR135 pKa = 11.84ITGKK139 pKa = 9.72VRR141 pKa = 11.84PSSPKK146 pKa = 9.64EE147 pKa = 4.08SNSSSTRR154 pKa = 11.84PSWLRR159 pKa = 11.84LLVEE163 pKa = 4.35VGLVAAAWYY172 pKa = 9.66GVYY175 pKa = 10.2KK176 pKa = 10.41YY177 pKa = 10.44ISLSEE182 pKa = 4.18EE183 pKa = 3.5IVRR186 pKa = 11.84RR187 pKa = 11.84DD188 pKa = 3.49QAQTRR193 pKa = 11.84PDD195 pKa = 3.34HH196 pKa = 6.32MRR198 pKa = 11.84QCYY201 pKa = 8.61EE202 pKa = 3.76QVKK205 pKa = 10.36EE206 pKa = 4.36SLEE209 pKa = 3.97EE210 pKa = 3.73QNNRR214 pKa = 11.84FIHH217 pKa = 6.89DD218 pKa = 3.49RR219 pKa = 11.84MEE221 pKa = 4.18EE222 pKa = 4.11EE223 pKa = 3.98LTKK226 pKa = 10.93LVIEE230 pKa = 4.31PAEE233 pKa = 3.92IDD235 pKa = 3.49EE236 pKa = 5.12DD237 pKa = 3.96GKK239 pKa = 10.2EE240 pKa = 4.11VKK242 pKa = 10.46PEE244 pKa = 3.59VSQYY248 pKa = 11.01LVRR251 pKa = 11.84HH252 pKa = 5.82HH253 pKa = 6.64GKK255 pKa = 9.77FVRR258 pKa = 11.84ALVTMAKK265 pKa = 10.16NEE267 pKa = 3.98FAGVPKK273 pKa = 9.12PTEE276 pKa = 4.15ANQLAVWRR284 pKa = 11.84FLYY287 pKa = 9.84RR288 pKa = 11.84QCDD291 pKa = 3.44KK292 pKa = 11.25RR293 pKa = 11.84GVNPTDD299 pKa = 3.45TQKK302 pKa = 11.05SISAALPFVFLPSAYY317 pKa = 10.34DD318 pKa = 3.28QDD320 pKa = 3.67QAITMNCDD328 pKa = 3.21DD329 pKa = 5.01AKK331 pKa = 11.47EE332 pKa = 3.94MLQRR336 pKa = 11.84YY337 pKa = 9.43ADD339 pKa = 3.73TFRR342 pKa = 11.84YY343 pKa = 6.1TTPLQKK349 pKa = 10.41LVHH352 pKa = 6.18NPLMGKK358 pKa = 8.59HH359 pKa = 3.98WVAWARR365 pKa = 11.84SIFISDD371 pKa = 3.99PEE373 pKa = 4.25TGLRR377 pKa = 11.84FAKK380 pKa = 10.5
MM1 pKa = 7.6EE2 pKa = 5.19SFVSRR7 pKa = 11.84ISEE10 pKa = 4.09ALSSCDD16 pKa = 4.77ANLDD20 pKa = 3.61YY21 pKa = 11.57DD22 pKa = 4.65KK23 pKa = 11.03MLLILKK29 pKa = 8.7HH30 pKa = 6.65CPDD33 pKa = 4.83KK34 pKa = 11.1LLSWASGIEE43 pKa = 3.95GLQTHH48 pKa = 5.38QLKK51 pKa = 10.33VAITHH56 pKa = 5.75LVKK59 pKa = 10.29TEE61 pKa = 3.53FHH63 pKa = 5.84TCKK66 pKa = 10.39HH67 pKa = 6.11LLTTMSATTADD78 pKa = 4.18YY79 pKa = 10.78IKK81 pKa = 10.35MFMEE85 pKa = 4.69LSPEE89 pKa = 3.79QEE91 pKa = 4.3VIINTGGTVSWWPKK105 pKa = 9.94LIRR108 pKa = 11.84DD109 pKa = 3.47IPEE112 pKa = 3.81WFTDD116 pKa = 4.49KK117 pKa = 10.61IPHH120 pKa = 6.57YY121 pKa = 9.81ILQIVDD127 pKa = 5.75FILRR131 pKa = 11.84WVSRR135 pKa = 11.84ITGKK139 pKa = 9.72VRR141 pKa = 11.84PSSPKK146 pKa = 9.64EE147 pKa = 4.08SNSSSTRR154 pKa = 11.84PSWLRR159 pKa = 11.84LLVEE163 pKa = 4.35VGLVAAAWYY172 pKa = 9.66GVYY175 pKa = 10.2KK176 pKa = 10.41YY177 pKa = 10.44ISLSEE182 pKa = 4.18EE183 pKa = 3.5IVRR186 pKa = 11.84RR187 pKa = 11.84DD188 pKa = 3.49QAQTRR193 pKa = 11.84PDD195 pKa = 3.34HH196 pKa = 6.32MRR198 pKa = 11.84QCYY201 pKa = 8.61EE202 pKa = 3.76QVKK205 pKa = 10.36EE206 pKa = 4.36SLEE209 pKa = 3.97EE210 pKa = 3.73QNNRR214 pKa = 11.84FIHH217 pKa = 6.89DD218 pKa = 3.49RR219 pKa = 11.84MEE221 pKa = 4.18EE222 pKa = 4.11EE223 pKa = 3.98LTKK226 pKa = 10.93LVIEE230 pKa = 4.31PAEE233 pKa = 3.92IDD235 pKa = 3.49EE236 pKa = 5.12DD237 pKa = 3.96GKK239 pKa = 10.2EE240 pKa = 4.11VKK242 pKa = 10.46PEE244 pKa = 3.59VSQYY248 pKa = 11.01LVRR251 pKa = 11.84HH252 pKa = 5.82HH253 pKa = 6.64GKK255 pKa = 9.77FVRR258 pKa = 11.84ALVTMAKK265 pKa = 10.16NEE267 pKa = 3.98FAGVPKK273 pKa = 9.12PTEE276 pKa = 4.15ANQLAVWRR284 pKa = 11.84FLYY287 pKa = 9.84RR288 pKa = 11.84QCDD291 pKa = 3.44KK292 pKa = 11.25RR293 pKa = 11.84GVNPTDD299 pKa = 3.45TQKK302 pKa = 11.05SISAALPFVFLPSAYY317 pKa = 10.34DD318 pKa = 3.28QDD320 pKa = 3.67QAITMNCDD328 pKa = 3.21DD329 pKa = 5.01AKK331 pKa = 11.47EE332 pKa = 3.94MLQRR336 pKa = 11.84YY337 pKa = 9.43ADD339 pKa = 3.73TFRR342 pKa = 11.84YY343 pKa = 6.1TTPLQKK349 pKa = 10.41LVHH352 pKa = 6.18NPLMGKK358 pKa = 8.59HH359 pKa = 3.98WVAWARR365 pKa = 11.84SIFISDD371 pKa = 3.99PEE373 pKa = 4.25TGLRR377 pKa = 11.84FAKK380 pKa = 10.5
Molecular weight: 43.88 kDa
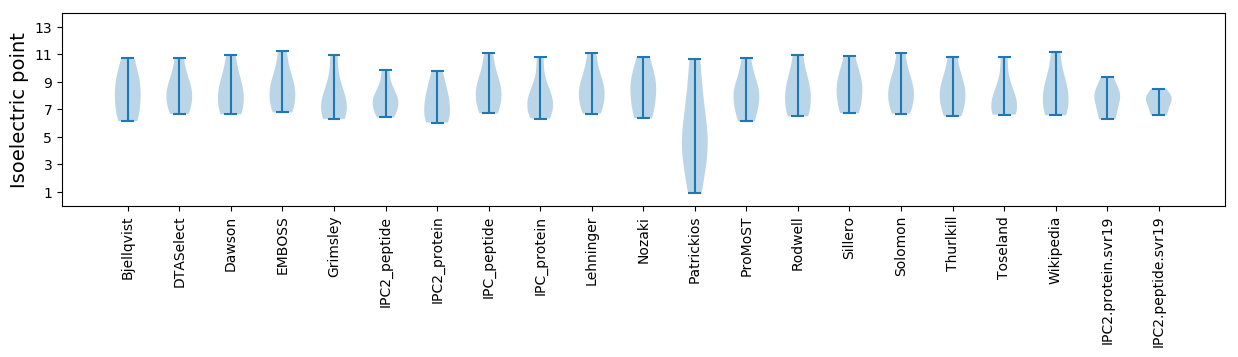
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S4WHA6|S4WHA6_9TOMB Capsid protein OS=Thin paspalum asymptomatic virus OX=1352511 PE=3 SV=1
MM1 pKa = 7.18NRR3 pKa = 11.84NNGAANARR11 pKa = 11.84RR12 pKa = 11.84GGKK15 pKa = 9.25RR16 pKa = 11.84APQSGARR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84ARR27 pKa = 11.84GKK29 pKa = 9.63SVEE32 pKa = 4.09RR33 pKa = 11.84RR34 pKa = 11.84AAPIQYY40 pKa = 7.99VTTVGPSQPRR50 pKa = 11.84MGTGQGWQRR59 pKa = 11.84ISHH62 pKa = 5.37QEE64 pKa = 3.6IILQVTASTTSDD76 pKa = 3.0SVQIVPIIPRR86 pKa = 11.84LSLSTAEE93 pKa = 3.99KK94 pKa = 10.16PIYY97 pKa = 9.69QGSAPHH103 pKa = 6.25LRR105 pKa = 11.84QMGDD109 pKa = 2.63AFSIHH114 pKa = 6.17RR115 pKa = 11.84WRR117 pKa = 11.84SLSFEE122 pKa = 4.41WVPSCPTTTPGNLVLRR138 pKa = 11.84FYY140 pKa = 10.52PSYY143 pKa = 10.13STPTPKK149 pKa = 10.8LLTDD153 pKa = 3.39IMDD156 pKa = 4.04SEE158 pKa = 4.87SLVIVPSISGAIYY171 pKa = 9.96RR172 pKa = 11.84PKK174 pKa = 10.11IDD176 pKa = 3.53TRR178 pKa = 11.84AQSPEE183 pKa = 3.83LRR185 pKa = 11.84NISITGFSALSDD197 pKa = 3.53EE198 pKa = 4.82DD199 pKa = 4.22KK200 pKa = 11.61GDD202 pKa = 3.7FSVGRR207 pKa = 11.84LVVGASKK214 pKa = 10.44QAVGLQLGLLRR225 pKa = 11.84MSYY228 pKa = 10.51VIEE231 pKa = 4.12LRR233 pKa = 11.84GATAGSGASAA243 pKa = 3.97
MM1 pKa = 7.18NRR3 pKa = 11.84NNGAANARR11 pKa = 11.84RR12 pKa = 11.84GGKK15 pKa = 9.25RR16 pKa = 11.84APQSGARR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84ARR27 pKa = 11.84GKK29 pKa = 9.63SVEE32 pKa = 4.09RR33 pKa = 11.84RR34 pKa = 11.84AAPIQYY40 pKa = 7.99VTTVGPSQPRR50 pKa = 11.84MGTGQGWQRR59 pKa = 11.84ISHH62 pKa = 5.37QEE64 pKa = 3.6IILQVTASTTSDD76 pKa = 3.0SVQIVPIIPRR86 pKa = 11.84LSLSTAEE93 pKa = 3.99KK94 pKa = 10.16PIYY97 pKa = 9.69QGSAPHH103 pKa = 6.25LRR105 pKa = 11.84QMGDD109 pKa = 2.63AFSIHH114 pKa = 6.17RR115 pKa = 11.84WRR117 pKa = 11.84SLSFEE122 pKa = 4.41WVPSCPTTTPGNLVLRR138 pKa = 11.84FYY140 pKa = 10.52PSYY143 pKa = 10.13STPTPKK149 pKa = 10.8LLTDD153 pKa = 3.39IMDD156 pKa = 4.04SEE158 pKa = 4.87SLVIVPSISGAIYY171 pKa = 9.96RR172 pKa = 11.84PKK174 pKa = 10.11IDD176 pKa = 3.53TRR178 pKa = 11.84AQSPEE183 pKa = 3.83LRR185 pKa = 11.84NISITGFSALSDD197 pKa = 3.53EE198 pKa = 4.82DD199 pKa = 4.22KK200 pKa = 11.61GDD202 pKa = 3.7FSVGRR207 pKa = 11.84LVVGASKK214 pKa = 10.44QAVGLQLGLLRR225 pKa = 11.84MSYY228 pKa = 10.51VIEE231 pKa = 4.12LRR233 pKa = 11.84GATAGSGASAA243 pKa = 3.97
Molecular weight: 26.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1834 |
59 |
947 |
305.7 |
34.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.143 ± 0.962 | 1.799 ± 0.37 |
4.689 ± 0.39 | 6.107 ± 0.65 |
3.381 ± 0.347 | 5.289 ± 0.795 |
2.944 ± 0.499 | 5.889 ± 0.375 |
6.107 ± 1.038 | 8.615 ± 0.489 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.454 ± 0.239 | 3.053 ± 0.436 |
6.107 ± 0.545 | 4.308 ± 0.364 |
6.707 ± 0.671 | 7.797 ± 0.98 |
5.671 ± 0.629 | 6.216 ± 0.328 |
1.963 ± 0.336 | 3.708 ± 0.577 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
