
Tomato torrado virus (isolate Solanum lycopersicum/Spain/PRIToTV0301/-) (ToTV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Picornavirales; Secoviridae; Torradovirus; Tomato torrado virus
Average proteome isoelectric point is 6.07
Get precalculated fractions of proteins
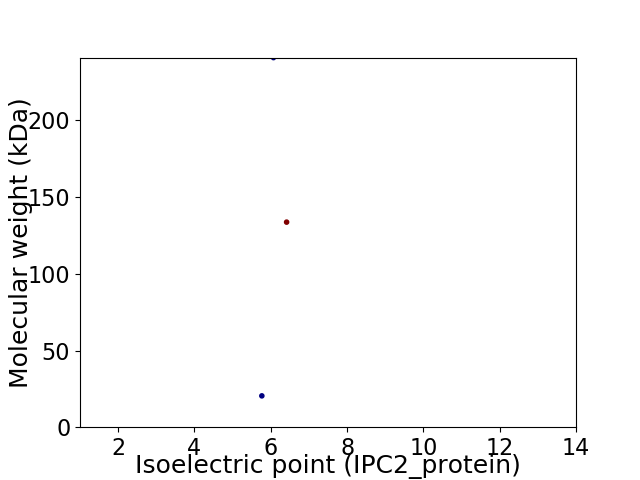
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
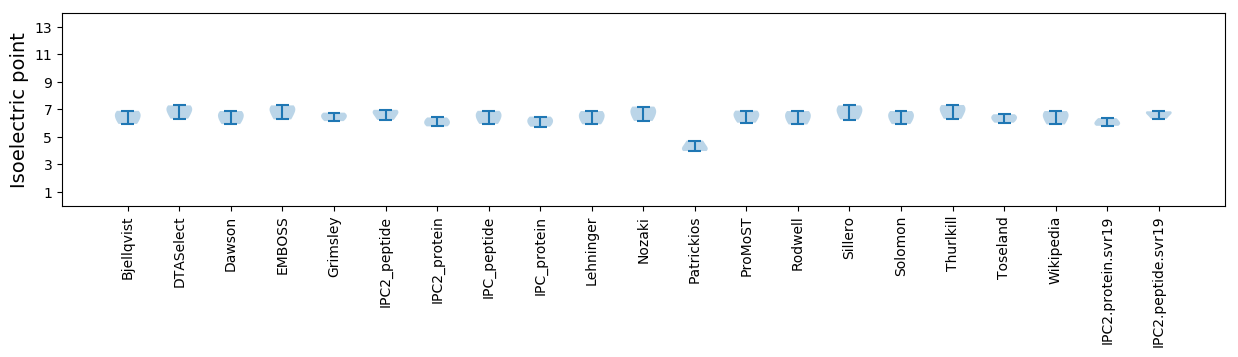
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|A1XIQ1|POL2_TOTV RNA2 polyprotein OS=Tomato torrado virus (isolate Solanum lycopersicum/Spain/PRIToTV0301/-) OX=686948 PE=3 SV=1
MM1 pKa = 7.64SFISRR6 pKa = 11.84LNTSLEE12 pKa = 3.75EE13 pKa = 4.06EE14 pKa = 4.39AFHH17 pKa = 6.82KK18 pKa = 10.6QVADD22 pKa = 4.0SQWVCSVDD30 pKa = 3.25TGSGIINSDD39 pKa = 3.12PTLDD43 pKa = 4.8FKK45 pKa = 11.03ICPKK49 pKa = 9.72TGGAISVLSVSWQNNSPQLVPGHH72 pKa = 5.74YY73 pKa = 10.32LLRR76 pKa = 11.84SGTWPITGVKK86 pKa = 10.32LSGLLVHH93 pKa = 6.98RR94 pKa = 11.84SIRR97 pKa = 11.84LEE99 pKa = 3.97TTRR102 pKa = 11.84KK103 pKa = 9.7LLEE106 pKa = 3.94AQRR109 pKa = 11.84ISVSQQASSSSAAGAAGKK127 pKa = 9.63QPQVTLTQLQEE138 pKa = 4.24EE139 pKa = 4.37LDD141 pKa = 3.82EE142 pKa = 4.85AKK144 pKa = 9.99TRR146 pKa = 11.84LALKK150 pKa = 10.18EE151 pKa = 4.09KK152 pKa = 10.17EE153 pKa = 4.0LLEE156 pKa = 4.25ALSEE160 pKa = 4.07ISKK163 pKa = 10.49LRR165 pKa = 11.84LQLSNQLSNDD175 pKa = 3.46DD176 pKa = 4.28VFSGWTEE183 pKa = 3.98EE184 pKa = 4.54GPKK187 pKa = 10.35
MM1 pKa = 7.64SFISRR6 pKa = 11.84LNTSLEE12 pKa = 3.75EE13 pKa = 4.06EE14 pKa = 4.39AFHH17 pKa = 6.82KK18 pKa = 10.6QVADD22 pKa = 4.0SQWVCSVDD30 pKa = 3.25TGSGIINSDD39 pKa = 3.12PTLDD43 pKa = 4.8FKK45 pKa = 11.03ICPKK49 pKa = 9.72TGGAISVLSVSWQNNSPQLVPGHH72 pKa = 5.74YY73 pKa = 10.32LLRR76 pKa = 11.84SGTWPITGVKK86 pKa = 10.32LSGLLVHH93 pKa = 6.98RR94 pKa = 11.84SIRR97 pKa = 11.84LEE99 pKa = 3.97TTRR102 pKa = 11.84KK103 pKa = 9.7LLEE106 pKa = 3.94AQRR109 pKa = 11.84ISVSQQASSSSAAGAAGKK127 pKa = 9.63QPQVTLTQLQEE138 pKa = 4.24EE139 pKa = 4.37LDD141 pKa = 3.82EE142 pKa = 4.85AKK144 pKa = 9.99TRR146 pKa = 11.84LALKK150 pKa = 10.18EE151 pKa = 4.09KK152 pKa = 10.17EE153 pKa = 4.0LLEE156 pKa = 4.25ALSEE160 pKa = 4.07ISKK163 pKa = 10.49LRR165 pKa = 11.84LQLSNQLSNDD175 pKa = 3.46DD176 pKa = 4.28VFSGWTEE183 pKa = 3.98EE184 pKa = 4.54GPKK187 pKa = 10.35
Molecular weight: 20.43 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|A1XIQ1|POL2_TOTV RNA2 polyprotein OS=Tomato torrado virus (isolate Solanum lycopersicum/Spain/PRIToTV0301/-) OX=686948 PE=3 SV=1
MM1 pKa = 7.12MMSSVAGQRR10 pKa = 11.84KK11 pKa = 9.1GPSDD15 pKa = 3.09KK16 pKa = 10.86SAAVSEE22 pKa = 3.98QLIAQITAAVEE33 pKa = 3.99AGNKK37 pKa = 9.32NLLRR41 pKa = 11.84KK42 pKa = 9.96LGMGSYY48 pKa = 10.47GILYY52 pKa = 8.87GAQEE56 pKa = 4.3KK57 pKa = 10.65KK58 pKa = 10.83AMEE61 pKa = 4.54LFDD64 pKa = 5.18PEE66 pKa = 4.44DD67 pKa = 3.44VHH69 pKa = 8.04NITSLWSSFKK79 pKa = 10.15KK80 pKa = 9.3TFTSSRR86 pKa = 11.84DD87 pKa = 3.2HH88 pKa = 6.8GNLFFHH94 pKa = 7.28LYY96 pKa = 10.04GVMFFMVPHH105 pKa = 4.83VHH107 pKa = 6.43GGEE110 pKa = 4.17GSVKK114 pKa = 10.24ISLCSSNDD122 pKa = 2.95PTNPVLQEE130 pKa = 3.68KK131 pKa = 10.15VLYY134 pKa = 10.2FSGGAQAVLMSPTITLPFVKK154 pKa = 9.56RR155 pKa = 11.84GPMFYY160 pKa = 10.23YY161 pKa = 10.55TMEE164 pKa = 4.43CLGTRR169 pKa = 11.84AQIPCSVVAIWKK181 pKa = 9.13QKK183 pKa = 9.82IDD185 pKa = 3.53IRR187 pKa = 11.84SAIYY191 pKa = 10.25SKK193 pKa = 10.89QEE195 pKa = 3.73TMSWAIEE202 pKa = 3.99ALHH205 pKa = 6.55RR206 pKa = 11.84PQFFQDD212 pKa = 3.13RR213 pKa = 11.84QEE215 pKa = 3.88AAQYY219 pKa = 10.21ISSVYY224 pKa = 11.1SNATSSATDD233 pKa = 3.21SVLPFVGAQLGDD245 pKa = 3.32TKK247 pKa = 10.98MNVPSEE253 pKa = 3.75ARR255 pKa = 11.84MIRR258 pKa = 11.84SSSLRR263 pKa = 11.84VPMLKK268 pKa = 9.89VQSKK272 pKa = 9.38RR273 pKa = 11.84FSSMEE278 pKa = 3.73IPSTSTAHH286 pKa = 7.04LLGTTRR292 pKa = 11.84DD293 pKa = 3.36EE294 pKa = 4.25TVIQEE299 pKa = 3.82EE300 pKa = 4.16SRR302 pKa = 11.84YY303 pKa = 9.96EE304 pKa = 3.89EE305 pKa = 4.48EE306 pKa = 5.41GDD308 pKa = 3.94DD309 pKa = 4.4GVLFPVKK316 pKa = 10.02KK317 pKa = 10.48AQGLNYY323 pKa = 10.27SHH325 pKa = 6.83VWDD328 pKa = 3.8NLGIEE333 pKa = 4.34SFVDD337 pKa = 3.65VEE339 pKa = 5.49LPEE342 pKa = 4.37NWDD345 pKa = 3.64EE346 pKa = 4.4LSVRR350 pKa = 11.84QQVAAAMIAFANKK363 pKa = 9.43GVCLVPKK370 pKa = 10.28HH371 pKa = 6.09IINRR375 pKa = 11.84DD376 pKa = 2.71KK377 pKa = 11.54HH378 pKa = 6.46NIHH381 pKa = 6.85LEE383 pKa = 4.04NITEE387 pKa = 4.06HH388 pKa = 6.6NYY390 pKa = 10.54LVILEE395 pKa = 4.52RR396 pKa = 11.84YY397 pKa = 9.15GIVNAGSLARR407 pKa = 11.84TEE409 pKa = 3.44NWYY412 pKa = 10.65NLTLAQRR419 pKa = 11.84VEE421 pKa = 3.86EE422 pKa = 5.0LIYY425 pKa = 10.83QRR427 pKa = 11.84DD428 pKa = 3.32DD429 pKa = 3.48AYY431 pKa = 11.5FMFGDD436 pKa = 3.79NTNPYY441 pKa = 9.94PPFDD445 pKa = 4.79CYY447 pKa = 11.31DD448 pKa = 3.51GLTLKK453 pKa = 10.26VRR455 pKa = 11.84SEE457 pKa = 4.22LEE459 pKa = 3.9RR460 pKa = 11.84VAKK463 pKa = 9.72EE464 pKa = 3.49QARR467 pKa = 11.84QRR469 pKa = 11.84FYY471 pKa = 11.54KK472 pKa = 10.02EE473 pKa = 3.14AARR476 pKa = 11.84AQVKK480 pKa = 10.57NKK482 pKa = 9.22VAQTSVEE489 pKa = 4.49EE490 pKa = 4.17IPSTSFATKK499 pKa = 9.94VAMEE503 pKa = 4.4SGSVDD508 pKa = 3.11SMKK511 pKa = 10.38IAIQAEE517 pKa = 4.16AANEE521 pKa = 3.76AVRR524 pKa = 11.84PNEE527 pKa = 3.56VMFEE531 pKa = 4.38FGQEE535 pKa = 4.05MNNEE539 pKa = 4.01GATEE543 pKa = 4.5LEE545 pKa = 4.48LQQPACVASNSFFNVGVFEE564 pKa = 4.31FAWKK568 pKa = 10.15KK569 pKa = 9.54SSSVAAEE576 pKa = 4.18VLSLALPAALFGKK589 pKa = 9.62SKK591 pKa = 9.86EE592 pKa = 4.05MSMGSQMLRR601 pKa = 11.84YY602 pKa = 9.29YY603 pKa = 10.75DD604 pKa = 3.95AALIMYY610 pKa = 9.11KK611 pKa = 10.38VILYY615 pKa = 9.67ISGMGAISGQLALVWDD631 pKa = 4.2EE632 pKa = 4.6CNVLNRR638 pKa = 11.84KK639 pKa = 9.54KK640 pKa = 10.71EE641 pKa = 4.27FINIASLYY649 pKa = 8.92ASKK652 pKa = 10.63HH653 pKa = 5.25RR654 pKa = 11.84LVSASEE660 pKa = 3.86QSSGEE665 pKa = 3.98FCFTPTGIGKK675 pKa = 8.15FVPLDD680 pKa = 3.94PASGAYY686 pKa = 9.86DD687 pKa = 3.65LGSIRR692 pKa = 11.84VFVTHH697 pKa = 7.12PLASATEE704 pKa = 4.29LEE706 pKa = 4.88SIPCHH711 pKa = 5.76IHH713 pKa = 5.81LQCKK717 pKa = 9.26VLSTNIMQPPRR728 pKa = 11.84LRR730 pKa = 11.84AQAQFGMKK738 pKa = 9.24PDD740 pKa = 3.44QTHH743 pKa = 6.61FPRR746 pKa = 11.84FPTNQVLLHH755 pKa = 5.74YY756 pKa = 9.13NWGVAASMGTTLVSIFSPSGIYY778 pKa = 10.36EE779 pKa = 4.06SDD781 pKa = 3.34GTLQPSLLGNIARR794 pKa = 11.84NCKK797 pKa = 7.76WWTGTCVFEE806 pKa = 4.01ICIEE810 pKa = 3.97KK811 pKa = 8.88TQFHH815 pKa = 6.65SGSLAIGLGTLNTSMSTPHH834 pKa = 7.59DD835 pKa = 4.16ILNMPHH841 pKa = 6.6VICNLEE847 pKa = 4.02MGRR850 pKa = 11.84KK851 pKa = 9.21FYY853 pKa = 10.6FRR855 pKa = 11.84CTITNWNGKK864 pKa = 8.59NLLTTGRR871 pKa = 11.84KK872 pKa = 9.24SSLPRR877 pKa = 11.84PKK879 pKa = 10.42HH880 pKa = 4.95MSHH883 pKa = 6.04MRR885 pKa = 11.84LFATVLKK892 pKa = 9.58PLVSTSIHH900 pKa = 7.0LDD902 pKa = 3.5TVGVTVQLKK911 pKa = 10.41CIEE914 pKa = 4.29DD915 pKa = 3.82LVLGGTVSVKK925 pKa = 10.22PIYY928 pKa = 10.11GHH930 pKa = 4.34WTKK933 pKa = 11.02GKK935 pKa = 9.77NAVDD939 pKa = 3.9FLFSEE944 pKa = 4.46MDD946 pKa = 3.71LSQRR950 pKa = 11.84KK951 pKa = 8.97EE952 pKa = 3.59IEE954 pKa = 3.91KK955 pKa = 10.27LRR957 pKa = 11.84KK958 pKa = 9.68EE959 pKa = 4.09NVEE962 pKa = 4.08TFDD965 pKa = 4.49EE966 pKa = 4.6KK967 pKa = 11.2GKK969 pKa = 10.12KK970 pKa = 8.62QPQVQVPLRR979 pKa = 11.84DD980 pKa = 3.47KK981 pKa = 10.73FSYY984 pKa = 10.59GAVQYY989 pKa = 10.81FVMNWKK995 pKa = 10.4DD996 pKa = 3.61EE997 pKa = 4.0EE998 pKa = 4.25RR999 pKa = 11.84LLVLPCAPWSVRR1011 pKa = 11.84FPQGALVQEE1020 pKa = 5.5AITCPFIDD1028 pKa = 3.66WCSSFCYY1035 pKa = 9.79WSGSLEE1041 pKa = 3.84YY1042 pKa = 9.98TIIVHH1047 pKa = 7.06RR1048 pKa = 11.84VQTSNNIGGVLNITLDD1064 pKa = 3.38SSGYY1068 pKa = 9.15PFPLGISKK1076 pKa = 8.4GTYY1079 pKa = 8.45VVSAGGGAKK1088 pKa = 8.96WAFTYY1093 pKa = 10.65GMSDD1097 pKa = 3.29NIFSFVVHH1105 pKa = 7.33DD1106 pKa = 4.82DD1107 pKa = 3.32EE1108 pKa = 5.35FFPRR1112 pKa = 11.84RR1113 pKa = 11.84HH1114 pKa = 4.97TKK1116 pKa = 10.04ARR1118 pKa = 11.84AIDD1121 pKa = 4.01PNASRR1126 pKa = 11.84IMTLQDD1132 pKa = 3.0RR1133 pKa = 11.84LGNLIINLPAKK1144 pKa = 10.36DD1145 pKa = 4.06VISSLEE1151 pKa = 3.85ILVKK1155 pKa = 10.41PGPDD1159 pKa = 3.26FKK1161 pKa = 11.26LQLAQAPSANHH1172 pKa = 6.07EE1173 pKa = 4.17KK1174 pKa = 10.66HH1175 pKa = 6.42LGDD1178 pKa = 4.0MQTHH1182 pKa = 6.59TYY1184 pKa = 10.56LYY1186 pKa = 9.85TPDD1189 pKa = 3.83FSEE1192 pKa = 4.91LRR1194 pKa = 11.84SFEE1197 pKa = 4.05NN1198 pKa = 3.5
MM1 pKa = 7.12MMSSVAGQRR10 pKa = 11.84KK11 pKa = 9.1GPSDD15 pKa = 3.09KK16 pKa = 10.86SAAVSEE22 pKa = 3.98QLIAQITAAVEE33 pKa = 3.99AGNKK37 pKa = 9.32NLLRR41 pKa = 11.84KK42 pKa = 9.96LGMGSYY48 pKa = 10.47GILYY52 pKa = 8.87GAQEE56 pKa = 4.3KK57 pKa = 10.65KK58 pKa = 10.83AMEE61 pKa = 4.54LFDD64 pKa = 5.18PEE66 pKa = 4.44DD67 pKa = 3.44VHH69 pKa = 8.04NITSLWSSFKK79 pKa = 10.15KK80 pKa = 9.3TFTSSRR86 pKa = 11.84DD87 pKa = 3.2HH88 pKa = 6.8GNLFFHH94 pKa = 7.28LYY96 pKa = 10.04GVMFFMVPHH105 pKa = 4.83VHH107 pKa = 6.43GGEE110 pKa = 4.17GSVKK114 pKa = 10.24ISLCSSNDD122 pKa = 2.95PTNPVLQEE130 pKa = 3.68KK131 pKa = 10.15VLYY134 pKa = 10.2FSGGAQAVLMSPTITLPFVKK154 pKa = 9.56RR155 pKa = 11.84GPMFYY160 pKa = 10.23YY161 pKa = 10.55TMEE164 pKa = 4.43CLGTRR169 pKa = 11.84AQIPCSVVAIWKK181 pKa = 9.13QKK183 pKa = 9.82IDD185 pKa = 3.53IRR187 pKa = 11.84SAIYY191 pKa = 10.25SKK193 pKa = 10.89QEE195 pKa = 3.73TMSWAIEE202 pKa = 3.99ALHH205 pKa = 6.55RR206 pKa = 11.84PQFFQDD212 pKa = 3.13RR213 pKa = 11.84QEE215 pKa = 3.88AAQYY219 pKa = 10.21ISSVYY224 pKa = 11.1SNATSSATDD233 pKa = 3.21SVLPFVGAQLGDD245 pKa = 3.32TKK247 pKa = 10.98MNVPSEE253 pKa = 3.75ARR255 pKa = 11.84MIRR258 pKa = 11.84SSSLRR263 pKa = 11.84VPMLKK268 pKa = 9.89VQSKK272 pKa = 9.38RR273 pKa = 11.84FSSMEE278 pKa = 3.73IPSTSTAHH286 pKa = 7.04LLGTTRR292 pKa = 11.84DD293 pKa = 3.36EE294 pKa = 4.25TVIQEE299 pKa = 3.82EE300 pKa = 4.16SRR302 pKa = 11.84YY303 pKa = 9.96EE304 pKa = 3.89EE305 pKa = 4.48EE306 pKa = 5.41GDD308 pKa = 3.94DD309 pKa = 4.4GVLFPVKK316 pKa = 10.02KK317 pKa = 10.48AQGLNYY323 pKa = 10.27SHH325 pKa = 6.83VWDD328 pKa = 3.8NLGIEE333 pKa = 4.34SFVDD337 pKa = 3.65VEE339 pKa = 5.49LPEE342 pKa = 4.37NWDD345 pKa = 3.64EE346 pKa = 4.4LSVRR350 pKa = 11.84QQVAAAMIAFANKK363 pKa = 9.43GVCLVPKK370 pKa = 10.28HH371 pKa = 6.09IINRR375 pKa = 11.84DD376 pKa = 2.71KK377 pKa = 11.54HH378 pKa = 6.46NIHH381 pKa = 6.85LEE383 pKa = 4.04NITEE387 pKa = 4.06HH388 pKa = 6.6NYY390 pKa = 10.54LVILEE395 pKa = 4.52RR396 pKa = 11.84YY397 pKa = 9.15GIVNAGSLARR407 pKa = 11.84TEE409 pKa = 3.44NWYY412 pKa = 10.65NLTLAQRR419 pKa = 11.84VEE421 pKa = 3.86EE422 pKa = 5.0LIYY425 pKa = 10.83QRR427 pKa = 11.84DD428 pKa = 3.32DD429 pKa = 3.48AYY431 pKa = 11.5FMFGDD436 pKa = 3.79NTNPYY441 pKa = 9.94PPFDD445 pKa = 4.79CYY447 pKa = 11.31DD448 pKa = 3.51GLTLKK453 pKa = 10.26VRR455 pKa = 11.84SEE457 pKa = 4.22LEE459 pKa = 3.9RR460 pKa = 11.84VAKK463 pKa = 9.72EE464 pKa = 3.49QARR467 pKa = 11.84QRR469 pKa = 11.84FYY471 pKa = 11.54KK472 pKa = 10.02EE473 pKa = 3.14AARR476 pKa = 11.84AQVKK480 pKa = 10.57NKK482 pKa = 9.22VAQTSVEE489 pKa = 4.49EE490 pKa = 4.17IPSTSFATKK499 pKa = 9.94VAMEE503 pKa = 4.4SGSVDD508 pKa = 3.11SMKK511 pKa = 10.38IAIQAEE517 pKa = 4.16AANEE521 pKa = 3.76AVRR524 pKa = 11.84PNEE527 pKa = 3.56VMFEE531 pKa = 4.38FGQEE535 pKa = 4.05MNNEE539 pKa = 4.01GATEE543 pKa = 4.5LEE545 pKa = 4.48LQQPACVASNSFFNVGVFEE564 pKa = 4.31FAWKK568 pKa = 10.15KK569 pKa = 9.54SSSVAAEE576 pKa = 4.18VLSLALPAALFGKK589 pKa = 9.62SKK591 pKa = 9.86EE592 pKa = 4.05MSMGSQMLRR601 pKa = 11.84YY602 pKa = 9.29YY603 pKa = 10.75DD604 pKa = 3.95AALIMYY610 pKa = 9.11KK611 pKa = 10.38VILYY615 pKa = 9.67ISGMGAISGQLALVWDD631 pKa = 4.2EE632 pKa = 4.6CNVLNRR638 pKa = 11.84KK639 pKa = 9.54KK640 pKa = 10.71EE641 pKa = 4.27FINIASLYY649 pKa = 8.92ASKK652 pKa = 10.63HH653 pKa = 5.25RR654 pKa = 11.84LVSASEE660 pKa = 3.86QSSGEE665 pKa = 3.98FCFTPTGIGKK675 pKa = 8.15FVPLDD680 pKa = 3.94PASGAYY686 pKa = 9.86DD687 pKa = 3.65LGSIRR692 pKa = 11.84VFVTHH697 pKa = 7.12PLASATEE704 pKa = 4.29LEE706 pKa = 4.88SIPCHH711 pKa = 5.76IHH713 pKa = 5.81LQCKK717 pKa = 9.26VLSTNIMQPPRR728 pKa = 11.84LRR730 pKa = 11.84AQAQFGMKK738 pKa = 9.24PDD740 pKa = 3.44QTHH743 pKa = 6.61FPRR746 pKa = 11.84FPTNQVLLHH755 pKa = 5.74YY756 pKa = 9.13NWGVAASMGTTLVSIFSPSGIYY778 pKa = 10.36EE779 pKa = 4.06SDD781 pKa = 3.34GTLQPSLLGNIARR794 pKa = 11.84NCKK797 pKa = 7.76WWTGTCVFEE806 pKa = 4.01ICIEE810 pKa = 3.97KK811 pKa = 8.88TQFHH815 pKa = 6.65SGSLAIGLGTLNTSMSTPHH834 pKa = 7.59DD835 pKa = 4.16ILNMPHH841 pKa = 6.6VICNLEE847 pKa = 4.02MGRR850 pKa = 11.84KK851 pKa = 9.21FYY853 pKa = 10.6FRR855 pKa = 11.84CTITNWNGKK864 pKa = 8.59NLLTTGRR871 pKa = 11.84KK872 pKa = 9.24SSLPRR877 pKa = 11.84PKK879 pKa = 10.42HH880 pKa = 4.95MSHH883 pKa = 6.04MRR885 pKa = 11.84LFATVLKK892 pKa = 9.58PLVSTSIHH900 pKa = 7.0LDD902 pKa = 3.5TVGVTVQLKK911 pKa = 10.41CIEE914 pKa = 4.29DD915 pKa = 3.82LVLGGTVSVKK925 pKa = 10.22PIYY928 pKa = 10.11GHH930 pKa = 4.34WTKK933 pKa = 11.02GKK935 pKa = 9.77NAVDD939 pKa = 3.9FLFSEE944 pKa = 4.46MDD946 pKa = 3.71LSQRR950 pKa = 11.84KK951 pKa = 8.97EE952 pKa = 3.59IEE954 pKa = 3.91KK955 pKa = 10.27LRR957 pKa = 11.84KK958 pKa = 9.68EE959 pKa = 4.09NVEE962 pKa = 4.08TFDD965 pKa = 4.49EE966 pKa = 4.6KK967 pKa = 11.2GKK969 pKa = 10.12KK970 pKa = 8.62QPQVQVPLRR979 pKa = 11.84DD980 pKa = 3.47KK981 pKa = 10.73FSYY984 pKa = 10.59GAVQYY989 pKa = 10.81FVMNWKK995 pKa = 10.4DD996 pKa = 3.61EE997 pKa = 4.0EE998 pKa = 4.25RR999 pKa = 11.84LLVLPCAPWSVRR1011 pKa = 11.84FPQGALVQEE1020 pKa = 5.5AITCPFIDD1028 pKa = 3.66WCSSFCYY1035 pKa = 9.79WSGSLEE1041 pKa = 3.84YY1042 pKa = 9.98TIIVHH1047 pKa = 7.06RR1048 pKa = 11.84VQTSNNIGGVLNITLDD1064 pKa = 3.38SSGYY1068 pKa = 9.15PFPLGISKK1076 pKa = 8.4GTYY1079 pKa = 8.45VVSAGGGAKK1088 pKa = 8.96WAFTYY1093 pKa = 10.65GMSDD1097 pKa = 3.29NIFSFVVHH1105 pKa = 7.33DD1106 pKa = 4.82DD1107 pKa = 3.32EE1108 pKa = 5.35FFPRR1112 pKa = 11.84RR1113 pKa = 11.84HH1114 pKa = 4.97TKK1116 pKa = 10.04ARR1118 pKa = 11.84AIDD1121 pKa = 4.01PNASRR1126 pKa = 11.84IMTLQDD1132 pKa = 3.0RR1133 pKa = 11.84LGNLIINLPAKK1144 pKa = 10.36DD1145 pKa = 4.06VISSLEE1151 pKa = 3.85ILVKK1155 pKa = 10.41PGPDD1159 pKa = 3.26FKK1161 pKa = 11.26LQLAQAPSANHH1172 pKa = 6.07EE1173 pKa = 4.17KK1174 pKa = 10.66HH1175 pKa = 6.42LGDD1178 pKa = 4.0MQTHH1182 pKa = 6.59TYY1184 pKa = 10.56LYY1186 pKa = 9.85TPDD1189 pKa = 3.83FSEE1192 pKa = 4.91LRR1194 pKa = 11.84SFEE1197 pKa = 4.05NN1198 pKa = 3.5
Molecular weight: 133.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3543 |
187 |
2158 |
1181.0 |
131.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.79 ± 0.404 | 2.06 ± 0.322 |
4.968 ± 0.677 | 5.391 ± 0.627 |
5.222 ± 0.63 | 6.068 ± 0.182 |
2.427 ± 0.159 | 5.476 ± 0.123 |
5.504 ± 0.073 | 9.173 ± 0.782 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.738 ± 0.443 | 4.036 ± 0.267 |
4.516 ± 0.171 | 4.29 ± 0.533 |
4.347 ± 0.054 | 8.326 ± 1.1 |
5.504 ± 0.169 | 7.423 ± 0.355 |
1.411 ± 0.16 | 3.302 ± 0.552 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
