
Leptidea sinapis
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Panarthropoda; Arthropoda; Mandibulata; Pancrustacea; Hexapoda; Insecta; Dicondylia; Pterygota; Neoptera; Endopterygota; Amphiesmenoptera; Lepidoptera; Glossata; Neolepidoptera; Heteroneura; Ditrysia;
Average proteome isoelectric point is 6.95
Get precalculated fractions of proteins
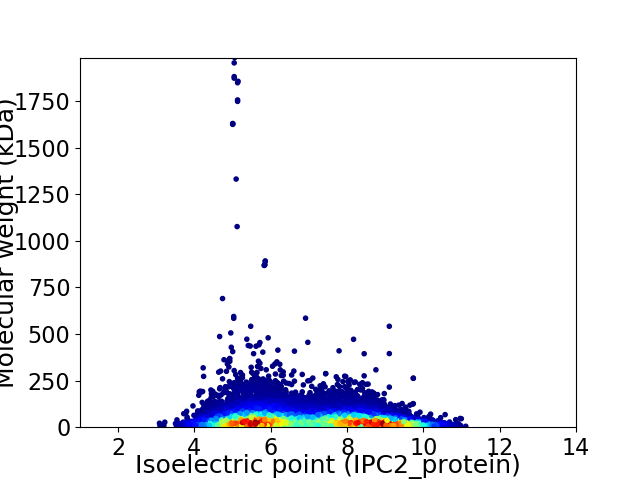
Virtual 2D-PAGE plot for 19862 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5E4R4H1|A0A5E4R4H1_9NEOP Uncharacterized protein OS=Leptidea sinapis OX=189913 GN=LSINAPIS_LOCUS14834 PE=4 SV=1
MM1 pKa = 7.32SRR3 pKa = 11.84NAKK6 pKa = 10.23AFGEE10 pKa = 4.51GEE12 pKa = 4.14PGDD15 pKa = 3.83VGDD18 pKa = 5.05RR19 pKa = 11.84DD20 pKa = 3.54VDD22 pKa = 3.95GAGPAATSTTTWRR35 pKa = 11.84QALTTDD41 pKa = 3.77GSGFTAKK48 pKa = 10.13PWPCCLDD55 pKa = 3.3ARR57 pKa = 11.84EE58 pKa = 4.25VPLAGQLNHH67 pKa = 6.21EE68 pKa = 4.67LAWGQGWGWVTLPNGVWSLTWSQDD92 pKa = 2.76GDD94 pKa = 4.21DD95 pKa = 4.43FALHH99 pKa = 6.97AMPIMHH105 pKa = 6.81HH106 pKa = 6.02TNVSQACSILDD117 pKa = 3.4GVTAYY122 pKa = 8.47EE123 pKa = 3.89ASAYY127 pKa = 8.61EE128 pKa = 4.02QALAEE133 pKa = 4.32YY134 pKa = 10.05DD135 pKa = 3.8DD136 pKa = 4.23TATTSDD142 pKa = 3.55DD143 pKa = 2.89WDD145 pKa = 4.32FGVDD149 pKa = 3.74CGDD152 pKa = 3.51FMMM155 pKa = 6.18
MM1 pKa = 7.32SRR3 pKa = 11.84NAKK6 pKa = 10.23AFGEE10 pKa = 4.51GEE12 pKa = 4.14PGDD15 pKa = 3.83VGDD18 pKa = 5.05RR19 pKa = 11.84DD20 pKa = 3.54VDD22 pKa = 3.95GAGPAATSTTTWRR35 pKa = 11.84QALTTDD41 pKa = 3.77GSGFTAKK48 pKa = 10.13PWPCCLDD55 pKa = 3.3ARR57 pKa = 11.84EE58 pKa = 4.25VPLAGQLNHH67 pKa = 6.21EE68 pKa = 4.67LAWGQGWGWVTLPNGVWSLTWSQDD92 pKa = 2.76GDD94 pKa = 4.21DD95 pKa = 4.43FALHH99 pKa = 6.97AMPIMHH105 pKa = 6.81HH106 pKa = 6.02TNVSQACSILDD117 pKa = 3.4GVTAYY122 pKa = 8.47EE123 pKa = 3.89ASAYY127 pKa = 8.61EE128 pKa = 4.02QALAEE133 pKa = 4.32YY134 pKa = 10.05DD135 pKa = 3.8DD136 pKa = 4.23TATTSDD142 pKa = 3.55DD143 pKa = 2.89WDD145 pKa = 4.32FGVDD149 pKa = 3.74CGDD152 pKa = 3.51FMMM155 pKa = 6.18
Molecular weight: 16.67 kDa
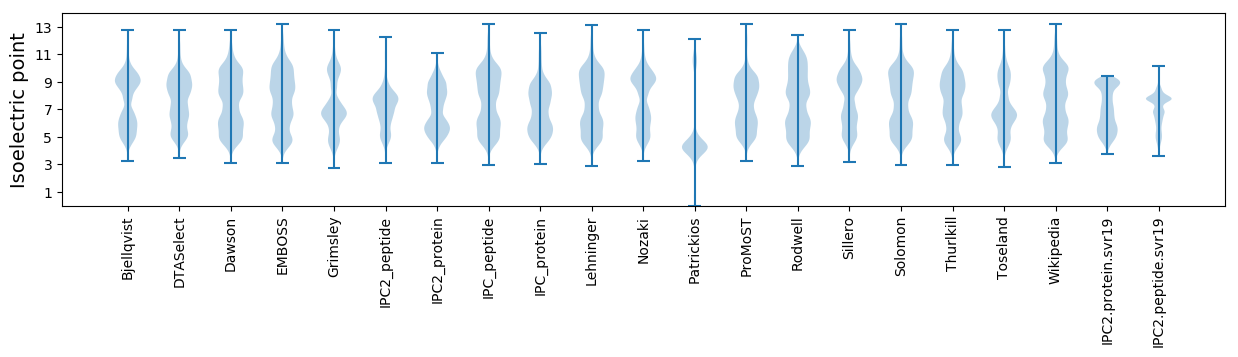
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5E4R0L8|A0A5E4R0L8_9NEOP Isoform of A0A5E4R152 Uncharacterized protein OS=Leptidea sinapis OX=189913 GN=LSINAPIS_LOCUS13011 PE=4 SV=1
MM1 pKa = 7.97DD2 pKa = 4.19VDD4 pKa = 3.59TDD6 pKa = 3.48GRR8 pKa = 11.84TSRR11 pKa = 11.84SRR13 pKa = 11.84SRR15 pKa = 11.84SVEE18 pKa = 3.77SKK20 pKa = 9.73MSHH23 pKa = 7.02DD24 pKa = 3.75GSKK27 pKa = 10.31SRR29 pKa = 11.84SRR31 pKa = 11.84SRR33 pKa = 11.84SGSKK37 pKa = 9.87SSSRR41 pKa = 11.84SRR43 pKa = 11.84SGSRR47 pKa = 11.84SSRR50 pKa = 11.84SRR52 pKa = 11.84SGSARR57 pKa = 11.84SRR59 pKa = 11.84SGSPQAISGGSPRR72 pKa = 11.84KK73 pKa = 9.44SRR75 pKa = 11.84SRR77 pKa = 11.84SSSRR81 pKa = 11.84RR82 pKa = 11.84SRR84 pKa = 11.84SGSASARR91 pKa = 11.84SRR93 pKa = 11.84SGSASVKK100 pKa = 9.91SRR102 pKa = 11.84SGSRR106 pKa = 11.84SRR108 pKa = 11.84SRR110 pKa = 11.84SRR112 pKa = 11.84SRR114 pKa = 11.84SGSVSPSRR122 pKa = 11.84ARR124 pKa = 11.84SRR126 pKa = 11.84SSNKK130 pKa = 9.98SVGKK134 pKa = 9.92SSKK137 pKa = 10.07NNSPARR143 pKa = 11.84SVSPANRR150 pKa = 11.84SGSVEE155 pKa = 4.0KK156 pKa = 10.21MDD158 pKa = 4.51VDD160 pKa = 3.84AEE162 pKa = 4.23EE163 pKa = 4.48NAPSRR168 pKa = 11.84TGSRR172 pKa = 11.84ASRR175 pKa = 11.84SRR177 pKa = 11.84SVSGSRR183 pKa = 11.84SRR185 pKa = 11.84SGSKK189 pKa = 10.38HH190 pKa = 5.67NSPEE194 pKa = 3.61DD195 pKa = 3.45ARR197 pKa = 11.84ASRR200 pKa = 11.84SRR202 pKa = 11.84SKK204 pKa = 10.66SGSRR208 pKa = 11.84SPVKK212 pKa = 10.12RR213 pKa = 11.84SKK215 pKa = 10.75SRR217 pKa = 11.84SKK219 pKa = 10.55SRR221 pKa = 11.84SRR223 pKa = 11.84SKK225 pKa = 10.6SRR227 pKa = 11.84SRR229 pKa = 11.84SKK231 pKa = 10.56SGSRR235 pKa = 11.84SKK237 pKa = 10.84SRR239 pKa = 11.84SRR241 pKa = 11.84SKK243 pKa = 10.56SGSRR247 pKa = 11.84SKK249 pKa = 10.84SRR251 pKa = 11.84SRR253 pKa = 11.84SKK255 pKa = 10.56SGSRR259 pKa = 11.84SKK261 pKa = 10.84SRR263 pKa = 11.84SRR265 pKa = 11.84SKK267 pKa = 10.6SRR269 pKa = 11.84SRR271 pKa = 11.84SKK273 pKa = 10.56SGSRR277 pKa = 11.84SKK279 pKa = 10.86SRR281 pKa = 11.84SKK283 pKa = 10.81SRR285 pKa = 11.84SKK287 pKa = 10.63SGSRR291 pKa = 11.84SKK293 pKa = 10.84SRR295 pKa = 11.84SRR297 pKa = 11.84SKK299 pKa = 10.6SRR301 pKa = 11.84SRR303 pKa = 11.84SKK305 pKa = 10.6SRR307 pKa = 11.84SRR309 pKa = 11.84SKK311 pKa = 10.6SRR313 pKa = 11.84SRR315 pKa = 11.84SKK317 pKa = 10.6SRR319 pKa = 11.84SRR321 pKa = 11.84SKK323 pKa = 10.56SGSRR327 pKa = 11.84SKK329 pKa = 10.84SRR331 pKa = 11.84SRR333 pKa = 11.84SKK335 pKa = 10.6SRR337 pKa = 11.84SRR339 pKa = 11.84SKK341 pKa = 10.6SRR343 pKa = 11.84SRR345 pKa = 11.84SKK347 pKa = 10.81SRR349 pKa = 11.84SKK351 pKa = 10.44SASRR355 pKa = 11.84AKK357 pKa = 10.38SGSRR361 pKa = 11.84SKK363 pKa = 10.89SKK365 pKa = 10.39SVSRR369 pKa = 11.84SRR371 pKa = 11.84SRR373 pKa = 11.84SRR375 pKa = 11.84SRR377 pKa = 11.84LKK379 pKa = 10.29SRR381 pKa = 11.84SRR383 pKa = 11.84VGSRR387 pKa = 11.84SGSAKK392 pKa = 9.64SVSRR396 pKa = 11.84SRR398 pKa = 11.84SGSRR402 pKa = 11.84AGSQSPARR410 pKa = 11.84KK411 pKa = 9.75GEE413 pKa = 3.44WRR415 pKa = 11.84AACRR419 pKa = 11.84TSRR422 pKa = 3.68
MM1 pKa = 7.97DD2 pKa = 4.19VDD4 pKa = 3.59TDD6 pKa = 3.48GRR8 pKa = 11.84TSRR11 pKa = 11.84SRR13 pKa = 11.84SRR15 pKa = 11.84SVEE18 pKa = 3.77SKK20 pKa = 9.73MSHH23 pKa = 7.02DD24 pKa = 3.75GSKK27 pKa = 10.31SRR29 pKa = 11.84SRR31 pKa = 11.84SRR33 pKa = 11.84SGSKK37 pKa = 9.87SSSRR41 pKa = 11.84SRR43 pKa = 11.84SGSRR47 pKa = 11.84SSRR50 pKa = 11.84SRR52 pKa = 11.84SGSARR57 pKa = 11.84SRR59 pKa = 11.84SGSPQAISGGSPRR72 pKa = 11.84KK73 pKa = 9.44SRR75 pKa = 11.84SRR77 pKa = 11.84SSSRR81 pKa = 11.84RR82 pKa = 11.84SRR84 pKa = 11.84SGSASARR91 pKa = 11.84SRR93 pKa = 11.84SGSASVKK100 pKa = 9.91SRR102 pKa = 11.84SGSRR106 pKa = 11.84SRR108 pKa = 11.84SRR110 pKa = 11.84SRR112 pKa = 11.84SRR114 pKa = 11.84SGSVSPSRR122 pKa = 11.84ARR124 pKa = 11.84SRR126 pKa = 11.84SSNKK130 pKa = 9.98SVGKK134 pKa = 9.92SSKK137 pKa = 10.07NNSPARR143 pKa = 11.84SVSPANRR150 pKa = 11.84SGSVEE155 pKa = 4.0KK156 pKa = 10.21MDD158 pKa = 4.51VDD160 pKa = 3.84AEE162 pKa = 4.23EE163 pKa = 4.48NAPSRR168 pKa = 11.84TGSRR172 pKa = 11.84ASRR175 pKa = 11.84SRR177 pKa = 11.84SVSGSRR183 pKa = 11.84SRR185 pKa = 11.84SGSKK189 pKa = 10.38HH190 pKa = 5.67NSPEE194 pKa = 3.61DD195 pKa = 3.45ARR197 pKa = 11.84ASRR200 pKa = 11.84SRR202 pKa = 11.84SKK204 pKa = 10.66SGSRR208 pKa = 11.84SPVKK212 pKa = 10.12RR213 pKa = 11.84SKK215 pKa = 10.75SRR217 pKa = 11.84SKK219 pKa = 10.55SRR221 pKa = 11.84SRR223 pKa = 11.84SKK225 pKa = 10.6SRR227 pKa = 11.84SRR229 pKa = 11.84SKK231 pKa = 10.56SGSRR235 pKa = 11.84SKK237 pKa = 10.84SRR239 pKa = 11.84SRR241 pKa = 11.84SKK243 pKa = 10.56SGSRR247 pKa = 11.84SKK249 pKa = 10.84SRR251 pKa = 11.84SRR253 pKa = 11.84SKK255 pKa = 10.56SGSRR259 pKa = 11.84SKK261 pKa = 10.84SRR263 pKa = 11.84SRR265 pKa = 11.84SKK267 pKa = 10.6SRR269 pKa = 11.84SRR271 pKa = 11.84SKK273 pKa = 10.56SGSRR277 pKa = 11.84SKK279 pKa = 10.86SRR281 pKa = 11.84SKK283 pKa = 10.81SRR285 pKa = 11.84SKK287 pKa = 10.63SGSRR291 pKa = 11.84SKK293 pKa = 10.84SRR295 pKa = 11.84SRR297 pKa = 11.84SKK299 pKa = 10.6SRR301 pKa = 11.84SRR303 pKa = 11.84SKK305 pKa = 10.6SRR307 pKa = 11.84SRR309 pKa = 11.84SKK311 pKa = 10.6SRR313 pKa = 11.84SRR315 pKa = 11.84SKK317 pKa = 10.6SRR319 pKa = 11.84SRR321 pKa = 11.84SKK323 pKa = 10.56SGSRR327 pKa = 11.84SKK329 pKa = 10.84SRR331 pKa = 11.84SRR333 pKa = 11.84SKK335 pKa = 10.6SRR337 pKa = 11.84SRR339 pKa = 11.84SKK341 pKa = 10.6SRR343 pKa = 11.84SRR345 pKa = 11.84SKK347 pKa = 10.81SRR349 pKa = 11.84SKK351 pKa = 10.44SASRR355 pKa = 11.84AKK357 pKa = 10.38SGSRR361 pKa = 11.84SKK363 pKa = 10.89SKK365 pKa = 10.39SVSRR369 pKa = 11.84SRR371 pKa = 11.84SRR373 pKa = 11.84SRR375 pKa = 11.84SRR377 pKa = 11.84LKK379 pKa = 10.29SRR381 pKa = 11.84SRR383 pKa = 11.84VGSRR387 pKa = 11.84SGSAKK392 pKa = 9.64SVSRR396 pKa = 11.84SRR398 pKa = 11.84SGSRR402 pKa = 11.84AGSQSPARR410 pKa = 11.84KK411 pKa = 9.75GEE413 pKa = 3.44WRR415 pKa = 11.84AACRR419 pKa = 11.84TSRR422 pKa = 3.68
Molecular weight: 45.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
7582099 |
8 |
18544 |
381.7 |
42.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.519 ± 0.024 | 2.27 ± 0.088 |
5.526 ± 0.019 | 6.579 ± 0.032 |
3.611 ± 0.019 | 5.467 ± 0.028 |
2.468 ± 0.011 | 5.711 ± 0.019 |
6.459 ± 0.036 | 9.013 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.264 ± 0.013 | 5.092 ± 0.025 |
5.25 ± 0.039 | 3.906 ± 0.018 |
5.621 ± 0.023 | 7.847 ± 0.023 |
5.721 ± 0.018 | 6.305 ± 0.017 |
1.111 ± 0.01 | 3.252 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
