
Tautonia plasticadhaerens
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Planctomycetia; Isosphaerales; Isosphaeraceae; Tautonia
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
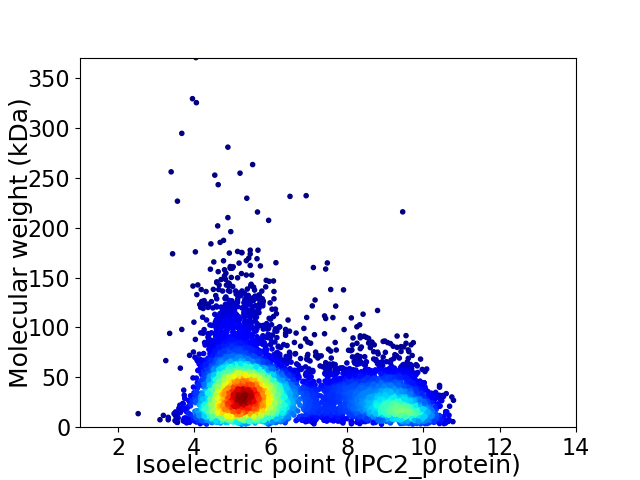
Virtual 2D-PAGE plot for 7326 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
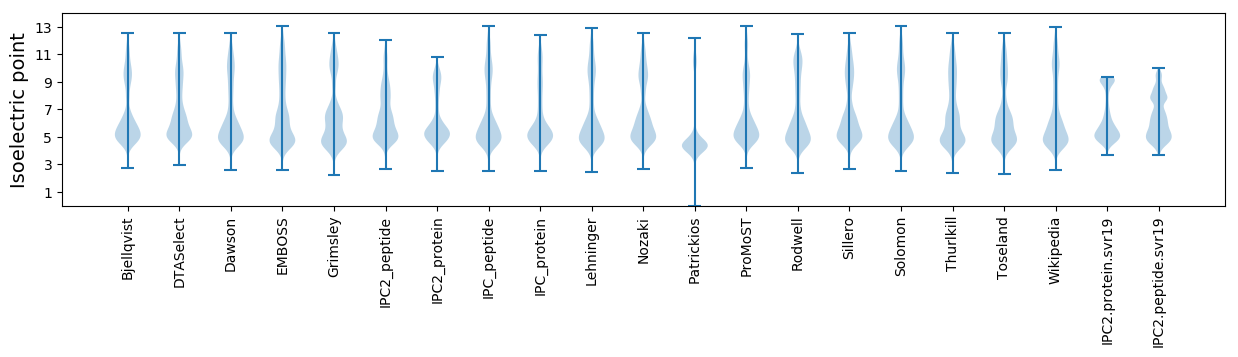
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A518HEX4|A0A518HEX4_9BACT Uncharacterized protein OS=Tautonia plasticadhaerens OX=2527974 GN=ElP_73740 PE=4 SV=1
MM1 pKa = 7.17SQFTHH6 pKa = 6.39YY7 pKa = 9.45EE8 pKa = 4.08HH9 pKa = 7.87PDD11 pKa = 3.38PASLEE16 pKa = 4.41DD17 pKa = 3.66YY18 pKa = 10.84CCICGWPLDD27 pKa = 4.03QPSLRR32 pKa = 11.84GLAATASPSGSGVGNDD48 pKa = 2.75WSYY51 pKa = 11.47FGQLHH56 pKa = 6.54LPSGLLSIADD66 pKa = 3.82PCEE69 pKa = 4.42CSDD72 pKa = 5.73DD73 pKa = 3.84GMALIDD79 pKa = 5.63LMPGDD84 pKa = 4.29YY85 pKa = 10.71AITIQTLLLDD95 pKa = 4.07RR96 pKa = 11.84ARR98 pKa = 11.84YY99 pKa = 4.97TARR102 pKa = 11.84VRR104 pKa = 11.84LASEE108 pKa = 4.2PDD110 pKa = 3.74GEE112 pKa = 5.03PGDD115 pKa = 4.81LLDD118 pKa = 5.71DD119 pKa = 3.7VWVDD123 pKa = 3.54GGQLLLFDD131 pKa = 6.26DD132 pKa = 5.06DD133 pKa = 5.44LGDD136 pKa = 3.91AAMTTVWTGFGDD148 pKa = 3.25VRR150 pKa = 11.84YY151 pKa = 9.24PIFEE155 pKa = 4.25VIKK158 pKa = 8.28EE159 pKa = 3.93GRR161 pKa = 11.84RR162 pKa = 11.84VGVEE166 pKa = 3.38IAFIDD171 pKa = 3.45IDD173 pKa = 4.09FPP175 pKa = 5.38
MM1 pKa = 7.17SQFTHH6 pKa = 6.39YY7 pKa = 9.45EE8 pKa = 4.08HH9 pKa = 7.87PDD11 pKa = 3.38PASLEE16 pKa = 4.41DD17 pKa = 3.66YY18 pKa = 10.84CCICGWPLDD27 pKa = 4.03QPSLRR32 pKa = 11.84GLAATASPSGSGVGNDD48 pKa = 2.75WSYY51 pKa = 11.47FGQLHH56 pKa = 6.54LPSGLLSIADD66 pKa = 3.82PCEE69 pKa = 4.42CSDD72 pKa = 5.73DD73 pKa = 3.84GMALIDD79 pKa = 5.63LMPGDD84 pKa = 4.29YY85 pKa = 10.71AITIQTLLLDD95 pKa = 4.07RR96 pKa = 11.84ARR98 pKa = 11.84YY99 pKa = 4.97TARR102 pKa = 11.84VRR104 pKa = 11.84LASEE108 pKa = 4.2PDD110 pKa = 3.74GEE112 pKa = 5.03PGDD115 pKa = 4.81LLDD118 pKa = 5.71DD119 pKa = 3.7VWVDD123 pKa = 3.54GGQLLLFDD131 pKa = 6.26DD132 pKa = 5.06DD133 pKa = 5.44LGDD136 pKa = 3.91AAMTTVWTGFGDD148 pKa = 3.25VRR150 pKa = 11.84YY151 pKa = 9.24PIFEE155 pKa = 4.25VIKK158 pKa = 8.28EE159 pKa = 3.93GRR161 pKa = 11.84RR162 pKa = 11.84VGVEE166 pKa = 3.38IAFIDD171 pKa = 3.45IDD173 pKa = 4.09FPP175 pKa = 5.38
Molecular weight: 19.09 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A518HFB3|A0A518HFB3_9BACT Non-specific serine/threonine protein kinase OS=Tautonia plasticadhaerens OX=2527974 GN=pknD_1 PE=4 SV=1
MM1 pKa = 7.7RR2 pKa = 11.84PGVAARR8 pKa = 11.84PDD10 pKa = 3.36PRR12 pKa = 11.84VARR15 pKa = 11.84GRR17 pKa = 11.84DD18 pKa = 3.31RR19 pKa = 11.84GPGPGSYY26 pKa = 10.19ARR28 pKa = 11.84SSARR32 pKa = 11.84IRR34 pKa = 11.84NRR36 pKa = 11.84RR37 pKa = 11.84SMPCLSSGRR46 pKa = 11.84TSSAGDD52 pKa = 3.1DD53 pKa = 3.17RR54 pKa = 11.84VRR56 pKa = 11.84RR57 pKa = 11.84EE58 pKa = 3.59PAGVGRR64 pKa = 11.84RR65 pKa = 11.84SASTGWSPGPAEE77 pKa = 4.5HH78 pKa = 7.01PHH80 pKa = 5.16RR81 pKa = 11.84HH82 pKa = 4.12QRR84 pKa = 11.84PRR86 pKa = 11.84LGAGLAPAGDD96 pKa = 3.38RR97 pKa = 11.84RR98 pKa = 11.84RR99 pKa = 11.84RR100 pKa = 11.84GRR102 pKa = 11.84RR103 pKa = 11.84HH104 pKa = 5.75DD105 pKa = 3.42RR106 pKa = 11.84LRR108 pKa = 11.84RR109 pKa = 11.84GARR112 pKa = 11.84RR113 pKa = 11.84PGHH116 pKa = 5.2RR117 pKa = 11.84AGRR120 pKa = 11.84RR121 pKa = 11.84RR122 pKa = 11.84AGPRR126 pKa = 11.84RR127 pKa = 11.84GPHH130 pKa = 6.05DD131 pKa = 3.78RR132 pKa = 11.84RR133 pKa = 11.84PGRR136 pKa = 11.84RR137 pKa = 11.84PAGRR141 pKa = 11.84RR142 pKa = 11.84RR143 pKa = 11.84GGPVSGLPGDD153 pKa = 3.88RR154 pKa = 11.84GRR156 pKa = 11.84RR157 pKa = 11.84GHH159 pKa = 7.06DD160 pKa = 3.28RR161 pKa = 11.84GPGDD165 pKa = 3.36HH166 pKa = 6.08QRR168 pKa = 11.84GRR170 pKa = 11.84RR171 pKa = 11.84GRR173 pKa = 11.84RR174 pKa = 11.84RR175 pKa = 11.84DD176 pKa = 3.07PRR178 pKa = 11.84RR179 pKa = 11.84RR180 pKa = 11.84GRR182 pKa = 11.84IDD184 pKa = 2.9ARR186 pKa = 11.84RR187 pKa = 11.84FAAGRR192 pKa = 11.84QPGRR196 pKa = 11.84RR197 pKa = 11.84RR198 pKa = 11.84RR199 pKa = 11.84ARR201 pKa = 11.84GRR203 pKa = 11.84RR204 pKa = 11.84PGRR207 pKa = 11.84GRR209 pKa = 11.84RR210 pKa = 11.84RR211 pKa = 11.84HH212 pKa = 4.59GRR214 pKa = 11.84GRR216 pKa = 11.84LAGRR220 pKa = 11.84RR221 pKa = 11.84HH222 pKa = 5.27EE223 pKa = 4.74HH224 pKa = 7.05LPLQPGRR231 pKa = 11.84GRR233 pKa = 11.84PGPGRR238 pKa = 11.84RR239 pKa = 11.84RR240 pKa = 3.36
MM1 pKa = 7.7RR2 pKa = 11.84PGVAARR8 pKa = 11.84PDD10 pKa = 3.36PRR12 pKa = 11.84VARR15 pKa = 11.84GRR17 pKa = 11.84DD18 pKa = 3.31RR19 pKa = 11.84GPGPGSYY26 pKa = 10.19ARR28 pKa = 11.84SSARR32 pKa = 11.84IRR34 pKa = 11.84NRR36 pKa = 11.84RR37 pKa = 11.84SMPCLSSGRR46 pKa = 11.84TSSAGDD52 pKa = 3.1DD53 pKa = 3.17RR54 pKa = 11.84VRR56 pKa = 11.84RR57 pKa = 11.84EE58 pKa = 3.59PAGVGRR64 pKa = 11.84RR65 pKa = 11.84SASTGWSPGPAEE77 pKa = 4.5HH78 pKa = 7.01PHH80 pKa = 5.16RR81 pKa = 11.84HH82 pKa = 4.12QRR84 pKa = 11.84PRR86 pKa = 11.84LGAGLAPAGDD96 pKa = 3.38RR97 pKa = 11.84RR98 pKa = 11.84RR99 pKa = 11.84RR100 pKa = 11.84GRR102 pKa = 11.84RR103 pKa = 11.84HH104 pKa = 5.75DD105 pKa = 3.42RR106 pKa = 11.84LRR108 pKa = 11.84RR109 pKa = 11.84GARR112 pKa = 11.84RR113 pKa = 11.84PGHH116 pKa = 5.2RR117 pKa = 11.84AGRR120 pKa = 11.84RR121 pKa = 11.84RR122 pKa = 11.84AGPRR126 pKa = 11.84RR127 pKa = 11.84GPHH130 pKa = 6.05DD131 pKa = 3.78RR132 pKa = 11.84RR133 pKa = 11.84PGRR136 pKa = 11.84RR137 pKa = 11.84PAGRR141 pKa = 11.84RR142 pKa = 11.84RR143 pKa = 11.84GGPVSGLPGDD153 pKa = 3.88RR154 pKa = 11.84GRR156 pKa = 11.84RR157 pKa = 11.84GHH159 pKa = 7.06DD160 pKa = 3.28RR161 pKa = 11.84GPGDD165 pKa = 3.36HH166 pKa = 6.08QRR168 pKa = 11.84GRR170 pKa = 11.84RR171 pKa = 11.84GRR173 pKa = 11.84RR174 pKa = 11.84RR175 pKa = 11.84DD176 pKa = 3.07PRR178 pKa = 11.84RR179 pKa = 11.84RR180 pKa = 11.84GRR182 pKa = 11.84IDD184 pKa = 2.9ARR186 pKa = 11.84RR187 pKa = 11.84FAAGRR192 pKa = 11.84QPGRR196 pKa = 11.84RR197 pKa = 11.84RR198 pKa = 11.84RR199 pKa = 11.84ARR201 pKa = 11.84GRR203 pKa = 11.84RR204 pKa = 11.84PGRR207 pKa = 11.84GRR209 pKa = 11.84RR210 pKa = 11.84RR211 pKa = 11.84HH212 pKa = 4.59GRR214 pKa = 11.84GRR216 pKa = 11.84LAGRR220 pKa = 11.84RR221 pKa = 11.84HH222 pKa = 5.27EE223 pKa = 4.74HH224 pKa = 7.05LPLQPGRR231 pKa = 11.84GRR233 pKa = 11.84PGPGRR238 pKa = 11.84RR239 pKa = 11.84RR240 pKa = 3.36
Molecular weight: 26.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2572131 |
29 |
3546 |
351.1 |
38.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.508 ± 0.039 | 0.884 ± 0.009 |
6.499 ± 0.024 | 6.313 ± 0.029 |
3.247 ± 0.015 | 9.765 ± 0.044 |
1.983 ± 0.016 | 3.962 ± 0.021 |
1.987 ± 0.021 | 10.263 ± 0.032 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.821 ± 0.012 | 1.946 ± 0.021 |
7.046 ± 0.032 | 2.5 ± 0.017 |
9.055 ± 0.039 | 5.561 ± 0.023 |
4.63 ± 0.028 | 7.416 ± 0.025 |
1.506 ± 0.014 | 2.108 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
