
Mycoplasma sp. (ex Biomphalaria glabrata)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Tenericutes; Mollicutes; Mycoplasmatales; Mycoplasmataceae; Mycoplasma; unclassified Mycoplasma
Average proteome isoelectric point is 7.2
Get precalculated fractions of proteins
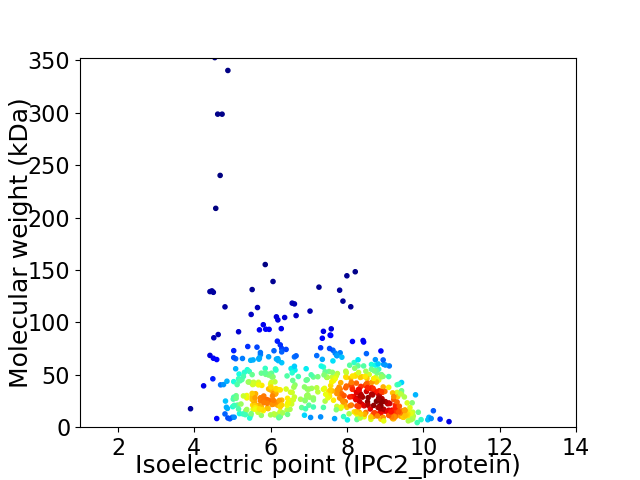
Virtual 2D-PAGE plot for 555 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0U3MRP5|A0A0U3MRP5_9MOLU YqgFc domain-containing protein OS=Mycoplasma sp. (ex Biomphalaria glabrata) OX=1749074 GN=ASO20_00455 PE=4 SV=1
MM1 pKa = 7.22NKK3 pKa = 9.66KK4 pKa = 10.13FKK6 pKa = 10.83LLALLATLTTVTLVPISSFMNRR28 pKa = 11.84NTNEE32 pKa = 3.61QQISKK37 pKa = 9.4NHH39 pKa = 5.27SQQTQTTSQTFNSTTQQVDD58 pKa = 3.7YY59 pKa = 10.33ASKK62 pKa = 10.61SIKK65 pKa = 9.9EE66 pKa = 3.96FNANVSPFLNQFEE79 pKa = 4.61FFLKK83 pKa = 10.55NGLYY87 pKa = 10.25FVDD90 pKa = 4.05LTSAYY95 pKa = 9.32EE96 pKa = 4.08NQATPSEE103 pKa = 4.26KK104 pKa = 10.53DD105 pKa = 3.47AIDD108 pKa = 5.8KK109 pKa = 10.51ISLWIGKK116 pKa = 9.78SSDD119 pKa = 3.3YY120 pKa = 10.72RR121 pKa = 11.84FSPMTNEE128 pKa = 3.93QLVAAATANFPEE140 pKa = 4.91DD141 pKa = 4.01LNPDD145 pKa = 3.48EE146 pKa = 5.03LATVIDD152 pKa = 4.31NFASIQSIAAKK163 pKa = 9.33TLIDD167 pKa = 3.56LTYY170 pKa = 10.61SKK172 pKa = 9.91WIQTMEE178 pKa = 4.02GTEE181 pKa = 5.63DD182 pKa = 3.05GHH184 pKa = 6.2QWGWYY189 pKa = 7.24NQNNLSNMFPAYY201 pKa = 10.69VIDD204 pKa = 4.15SKK206 pKa = 11.81NGLTFDD212 pKa = 4.69NKK214 pKa = 10.8DD215 pKa = 3.77DD216 pKa = 5.08NIANPGVGLQNLLAKK231 pKa = 10.6FHH233 pKa = 7.5DD234 pKa = 4.27SLEE237 pKa = 4.37AGNPEE242 pKa = 4.73RR243 pKa = 11.84LTEE246 pKa = 4.43PYY248 pKa = 10.06LNPVSGSAIEE258 pKa = 4.33TAFHH262 pKa = 7.66EE263 pKa = 4.46YY264 pKa = 8.07QTWGKK269 pKa = 8.85NQFSYY274 pKa = 11.06YY275 pKa = 7.43VQKK278 pKa = 10.71IINQYY283 pKa = 9.33NSFYY287 pKa = 11.04YY288 pKa = 10.8GFVDD292 pKa = 5.76LNSQNQQYY300 pKa = 8.82WNYY303 pKa = 9.7YY304 pKa = 10.12LDD306 pKa = 5.09DD307 pKa = 3.88ATLNPLTSVDD317 pKa = 3.69EE318 pKa = 5.04VISKK322 pKa = 8.81WINVMKK328 pKa = 10.52DD329 pKa = 3.4PSWNASTVINFLFSTDD345 pKa = 2.58VWAGLSVDD353 pKa = 3.84SVGGKK358 pKa = 9.8DD359 pKa = 3.38ATIEE363 pKa = 3.92QYY365 pKa = 11.29EE366 pKa = 4.43FVATADD372 pKa = 3.81FNNFTLINTNSFKK385 pKa = 11.01LATNTLYY392 pKa = 11.11NQFLEE397 pKa = 4.24NNYY400 pKa = 10.1FGYY403 pKa = 10.97ADD405 pKa = 3.76PNGVFNVSQYY415 pKa = 10.9FSGTPTQIVTMQEE428 pKa = 4.08FKK430 pKa = 11.2DD431 pKa = 4.07SFIHH435 pKa = 5.86TAATYY440 pKa = 10.22IEE442 pKa = 4.07LSKK445 pKa = 11.13YY446 pKa = 9.68FIWTSEE452 pKa = 3.43GSYY455 pKa = 10.51YY456 pKa = 10.51RR457 pKa = 11.84PAEE460 pKa = 3.88NQNFLNYY467 pKa = 9.84VFYY470 pKa = 10.56QQSSLEE476 pKa = 3.86QDD478 pKa = 3.48FQSIIDD484 pKa = 4.15LVSPSYY490 pKa = 10.86YY491 pKa = 10.68GYY493 pKa = 11.45ASLANNRR500 pKa = 11.84LTTYY504 pKa = 10.16IFQNLEE510 pKa = 3.66VEE512 pKa = 4.59KK513 pKa = 10.98QMSAADD519 pKa = 3.6LADD522 pKa = 3.78YY523 pKa = 10.04FILNHH528 pKa = 6.82PATSDD533 pKa = 3.25EE534 pKa = 4.27YY535 pKa = 11.3LGVWATKK542 pKa = 11.08SMIDD546 pKa = 3.55SLKK549 pKa = 10.96SNPEE553 pKa = 3.68LGLNDD558 pKa = 3.52WVNDD562 pKa = 3.9QKK564 pKa = 9.85TQLNNDD570 pKa = 3.52LQKK573 pKa = 10.78IIDD576 pKa = 4.44KK577 pKa = 10.95IKK579 pKa = 10.46DD580 pKa = 3.44NYY582 pKa = 9.66FGWVTTTSGFGLLVSNVDD600 pKa = 3.28KK601 pKa = 11.21LLVNFPVNTEE611 pKa = 4.39LSPEE615 pKa = 4.49AIAATITMEE624 pKa = 4.21SLPTGLTGLYY634 pKa = 9.33EE635 pKa = 3.96IKK637 pKa = 10.82GFLKK641 pKa = 11.02LITSDD646 pKa = 5.5DD647 pKa = 3.45INTFKK652 pKa = 11.26NNEE655 pKa = 4.08SKK657 pKa = 11.08YY658 pKa = 10.06LSQEE662 pKa = 3.07ISRR665 pKa = 11.84IVPLFEE671 pKa = 3.56NTYY674 pKa = 8.89YY675 pKa = 10.83GYY677 pKa = 10.32SASPEE682 pKa = 3.86FGANAIYY689 pKa = 10.1QVIFYY694 pKa = 7.21THH696 pKa = 6.69RR697 pKa = 11.84TNTPPNEE704 pKa = 4.01QMIVDD709 pKa = 4.41AFKK712 pKa = 10.87QGLSVSAEE720 pKa = 4.16GFPKK724 pKa = 10.29PYY726 pKa = 10.21LVRR729 pKa = 11.84QFLGNITIEE738 pKa = 4.21NTDD741 pKa = 3.64FQAFVQDD748 pKa = 3.26QRR750 pKa = 11.84KK751 pKa = 8.21QVIDD755 pKa = 3.75HH756 pKa = 6.35FTTIIGMIEE765 pKa = 3.78PYY767 pKa = 10.4YY768 pKa = 10.84YY769 pKa = 10.93GFEE772 pKa = 3.83RR773 pKa = 11.84LANNSVSDD781 pKa = 4.08YY782 pKa = 11.46VLLGYY787 pKa = 9.2TDD789 pKa = 4.19GTPLDD794 pKa = 3.94PVVLANYY801 pKa = 10.58LMDD804 pKa = 4.79NFPSTPTIDD813 pKa = 5.92LPGKK817 pKa = 8.79WYY819 pKa = 11.06LNGFLTTLITDD830 pKa = 3.89KK831 pKa = 10.77TAINEE836 pKa = 4.06FLTDD840 pKa = 3.84QSNSLQKK847 pKa = 10.55AWNPIYY853 pKa = 10.78DD854 pKa = 3.66EE855 pKa = 4.42MVNNYY860 pKa = 9.72YY861 pKa = 10.88GYY863 pKa = 7.02TTEE866 pKa = 4.03MDD868 pKa = 3.71SLVSRR873 pKa = 11.84TLLIIQNDD881 pKa = 4.29GTPLDD886 pKa = 3.94SVALLKK892 pKa = 10.76LFKK895 pKa = 10.91QNVIDD900 pKa = 3.56MHH902 pKa = 6.05GTQDD906 pKa = 3.34FPFSYY911 pKa = 10.21GVKK914 pKa = 9.53VWLEE918 pKa = 3.82TLVNEE923 pKa = 4.42NKK925 pKa = 10.02ISGEE929 pKa = 3.92FKK931 pKa = 10.71DD932 pKa = 3.98EE933 pKa = 3.69QRR935 pKa = 11.84TALSYY940 pKa = 10.7DD941 pKa = 3.05IEE943 pKa = 4.52HH944 pKa = 7.41AVRR947 pKa = 11.84EE948 pKa = 4.26YY949 pKa = 11.43NNAYY953 pKa = 8.81WGYY956 pKa = 11.09NNDD959 pKa = 3.46TTLPNYY965 pKa = 10.15FIEE968 pKa = 4.44YY969 pKa = 10.4FLGSDD974 pKa = 3.52HH975 pKa = 6.61KK976 pKa = 9.92TPLTNAQIEE985 pKa = 4.77SNILDD990 pKa = 3.86AAKK993 pKa = 10.16GWGFYY998 pKa = 10.33KK999 pKa = 10.21IYY1001 pKa = 10.88DD1002 pKa = 3.93FFNSGISGGVFYY1014 pKa = 11.02DD1015 pKa = 3.37LFIFSEE1021 pKa = 4.28SQEE1024 pKa = 4.26VIAKK1028 pKa = 10.15GDD1030 pKa = 3.5LRR1032 pKa = 11.84NILEE1036 pKa = 5.07LLVQYY1041 pKa = 11.37YY1042 pKa = 10.33NDD1044 pKa = 3.49KK1045 pKa = 11.05DD1046 pKa = 3.81FNYY1049 pKa = 8.78TQTPSPFTGANHH1061 pKa = 6.48IWTLFICKK1069 pKa = 10.06NGSTYY1074 pKa = 11.07GLEE1077 pKa = 4.24SNVDD1081 pKa = 3.3GLYY1084 pKa = 10.19TLFKK1088 pKa = 10.78AQIAQTDD1095 pKa = 4.81LINRR1099 pKa = 11.84ITHH1102 pKa = 5.3YY1103 pKa = 10.86SVVAEE1108 pKa = 4.15YY1109 pKa = 10.84LKK1111 pKa = 10.84SLYY1114 pKa = 11.13VNDD1117 pKa = 4.78KK1118 pKa = 9.33GTLSAAGEE1126 pKa = 4.04LLNWKK1131 pKa = 8.55EE1132 pKa = 4.03WEE1134 pKa = 4.34GQQ1136 pKa = 3.33
MM1 pKa = 7.22NKK3 pKa = 9.66KK4 pKa = 10.13FKK6 pKa = 10.83LLALLATLTTVTLVPISSFMNRR28 pKa = 11.84NTNEE32 pKa = 3.61QQISKK37 pKa = 9.4NHH39 pKa = 5.27SQQTQTTSQTFNSTTQQVDD58 pKa = 3.7YY59 pKa = 10.33ASKK62 pKa = 10.61SIKK65 pKa = 9.9EE66 pKa = 3.96FNANVSPFLNQFEE79 pKa = 4.61FFLKK83 pKa = 10.55NGLYY87 pKa = 10.25FVDD90 pKa = 4.05LTSAYY95 pKa = 9.32EE96 pKa = 4.08NQATPSEE103 pKa = 4.26KK104 pKa = 10.53DD105 pKa = 3.47AIDD108 pKa = 5.8KK109 pKa = 10.51ISLWIGKK116 pKa = 9.78SSDD119 pKa = 3.3YY120 pKa = 10.72RR121 pKa = 11.84FSPMTNEE128 pKa = 3.93QLVAAATANFPEE140 pKa = 4.91DD141 pKa = 4.01LNPDD145 pKa = 3.48EE146 pKa = 5.03LATVIDD152 pKa = 4.31NFASIQSIAAKK163 pKa = 9.33TLIDD167 pKa = 3.56LTYY170 pKa = 10.61SKK172 pKa = 9.91WIQTMEE178 pKa = 4.02GTEE181 pKa = 5.63DD182 pKa = 3.05GHH184 pKa = 6.2QWGWYY189 pKa = 7.24NQNNLSNMFPAYY201 pKa = 10.69VIDD204 pKa = 4.15SKK206 pKa = 11.81NGLTFDD212 pKa = 4.69NKK214 pKa = 10.8DD215 pKa = 3.77DD216 pKa = 5.08NIANPGVGLQNLLAKK231 pKa = 10.6FHH233 pKa = 7.5DD234 pKa = 4.27SLEE237 pKa = 4.37AGNPEE242 pKa = 4.73RR243 pKa = 11.84LTEE246 pKa = 4.43PYY248 pKa = 10.06LNPVSGSAIEE258 pKa = 4.33TAFHH262 pKa = 7.66EE263 pKa = 4.46YY264 pKa = 8.07QTWGKK269 pKa = 8.85NQFSYY274 pKa = 11.06YY275 pKa = 7.43VQKK278 pKa = 10.71IINQYY283 pKa = 9.33NSFYY287 pKa = 11.04YY288 pKa = 10.8GFVDD292 pKa = 5.76LNSQNQQYY300 pKa = 8.82WNYY303 pKa = 9.7YY304 pKa = 10.12LDD306 pKa = 5.09DD307 pKa = 3.88ATLNPLTSVDD317 pKa = 3.69EE318 pKa = 5.04VISKK322 pKa = 8.81WINVMKK328 pKa = 10.52DD329 pKa = 3.4PSWNASTVINFLFSTDD345 pKa = 2.58VWAGLSVDD353 pKa = 3.84SVGGKK358 pKa = 9.8DD359 pKa = 3.38ATIEE363 pKa = 3.92QYY365 pKa = 11.29EE366 pKa = 4.43FVATADD372 pKa = 3.81FNNFTLINTNSFKK385 pKa = 11.01LATNTLYY392 pKa = 11.11NQFLEE397 pKa = 4.24NNYY400 pKa = 10.1FGYY403 pKa = 10.97ADD405 pKa = 3.76PNGVFNVSQYY415 pKa = 10.9FSGTPTQIVTMQEE428 pKa = 4.08FKK430 pKa = 11.2DD431 pKa = 4.07SFIHH435 pKa = 5.86TAATYY440 pKa = 10.22IEE442 pKa = 4.07LSKK445 pKa = 11.13YY446 pKa = 9.68FIWTSEE452 pKa = 3.43GSYY455 pKa = 10.51YY456 pKa = 10.51RR457 pKa = 11.84PAEE460 pKa = 3.88NQNFLNYY467 pKa = 9.84VFYY470 pKa = 10.56QQSSLEE476 pKa = 3.86QDD478 pKa = 3.48FQSIIDD484 pKa = 4.15LVSPSYY490 pKa = 10.86YY491 pKa = 10.68GYY493 pKa = 11.45ASLANNRR500 pKa = 11.84LTTYY504 pKa = 10.16IFQNLEE510 pKa = 3.66VEE512 pKa = 4.59KK513 pKa = 10.98QMSAADD519 pKa = 3.6LADD522 pKa = 3.78YY523 pKa = 10.04FILNHH528 pKa = 6.82PATSDD533 pKa = 3.25EE534 pKa = 4.27YY535 pKa = 11.3LGVWATKK542 pKa = 11.08SMIDD546 pKa = 3.55SLKK549 pKa = 10.96SNPEE553 pKa = 3.68LGLNDD558 pKa = 3.52WVNDD562 pKa = 3.9QKK564 pKa = 9.85TQLNNDD570 pKa = 3.52LQKK573 pKa = 10.78IIDD576 pKa = 4.44KK577 pKa = 10.95IKK579 pKa = 10.46DD580 pKa = 3.44NYY582 pKa = 9.66FGWVTTTSGFGLLVSNVDD600 pKa = 3.28KK601 pKa = 11.21LLVNFPVNTEE611 pKa = 4.39LSPEE615 pKa = 4.49AIAATITMEE624 pKa = 4.21SLPTGLTGLYY634 pKa = 9.33EE635 pKa = 3.96IKK637 pKa = 10.82GFLKK641 pKa = 11.02LITSDD646 pKa = 5.5DD647 pKa = 3.45INTFKK652 pKa = 11.26NNEE655 pKa = 4.08SKK657 pKa = 11.08YY658 pKa = 10.06LSQEE662 pKa = 3.07ISRR665 pKa = 11.84IVPLFEE671 pKa = 3.56NTYY674 pKa = 8.89YY675 pKa = 10.83GYY677 pKa = 10.32SASPEE682 pKa = 3.86FGANAIYY689 pKa = 10.1QVIFYY694 pKa = 7.21THH696 pKa = 6.69RR697 pKa = 11.84TNTPPNEE704 pKa = 4.01QMIVDD709 pKa = 4.41AFKK712 pKa = 10.87QGLSVSAEE720 pKa = 4.16GFPKK724 pKa = 10.29PYY726 pKa = 10.21LVRR729 pKa = 11.84QFLGNITIEE738 pKa = 4.21NTDD741 pKa = 3.64FQAFVQDD748 pKa = 3.26QRR750 pKa = 11.84KK751 pKa = 8.21QVIDD755 pKa = 3.75HH756 pKa = 6.35FTTIIGMIEE765 pKa = 3.78PYY767 pKa = 10.4YY768 pKa = 10.84YY769 pKa = 10.93GFEE772 pKa = 3.83RR773 pKa = 11.84LANNSVSDD781 pKa = 4.08YY782 pKa = 11.46VLLGYY787 pKa = 9.2TDD789 pKa = 4.19GTPLDD794 pKa = 3.94PVVLANYY801 pKa = 10.58LMDD804 pKa = 4.79NFPSTPTIDD813 pKa = 5.92LPGKK817 pKa = 8.79WYY819 pKa = 11.06LNGFLTTLITDD830 pKa = 3.89KK831 pKa = 10.77TAINEE836 pKa = 4.06FLTDD840 pKa = 3.84QSNSLQKK847 pKa = 10.55AWNPIYY853 pKa = 10.78DD854 pKa = 3.66EE855 pKa = 4.42MVNNYY860 pKa = 9.72YY861 pKa = 10.88GYY863 pKa = 7.02TTEE866 pKa = 4.03MDD868 pKa = 3.71SLVSRR873 pKa = 11.84TLLIIQNDD881 pKa = 4.29GTPLDD886 pKa = 3.94SVALLKK892 pKa = 10.76LFKK895 pKa = 10.91QNVIDD900 pKa = 3.56MHH902 pKa = 6.05GTQDD906 pKa = 3.34FPFSYY911 pKa = 10.21GVKK914 pKa = 9.53VWLEE918 pKa = 3.82TLVNEE923 pKa = 4.42NKK925 pKa = 10.02ISGEE929 pKa = 3.92FKK931 pKa = 10.71DD932 pKa = 3.98EE933 pKa = 3.69QRR935 pKa = 11.84TALSYY940 pKa = 10.7DD941 pKa = 3.05IEE943 pKa = 4.52HH944 pKa = 7.41AVRR947 pKa = 11.84EE948 pKa = 4.26YY949 pKa = 11.43NNAYY953 pKa = 8.81WGYY956 pKa = 11.09NNDD959 pKa = 3.46TTLPNYY965 pKa = 10.15FIEE968 pKa = 4.44YY969 pKa = 10.4FLGSDD974 pKa = 3.52HH975 pKa = 6.61KK976 pKa = 9.92TPLTNAQIEE985 pKa = 4.77SNILDD990 pKa = 3.86AAKK993 pKa = 10.16GWGFYY998 pKa = 10.33KK999 pKa = 10.21IYY1001 pKa = 10.88DD1002 pKa = 3.93FFNSGISGGVFYY1014 pKa = 11.02DD1015 pKa = 3.37LFIFSEE1021 pKa = 4.28SQEE1024 pKa = 4.26VIAKK1028 pKa = 10.15GDD1030 pKa = 3.5LRR1032 pKa = 11.84NILEE1036 pKa = 5.07LLVQYY1041 pKa = 11.37YY1042 pKa = 10.33NDD1044 pKa = 3.49KK1045 pKa = 11.05DD1046 pKa = 3.81FNYY1049 pKa = 8.78TQTPSPFTGANHH1061 pKa = 6.48IWTLFICKK1069 pKa = 10.06NGSTYY1074 pKa = 11.07GLEE1077 pKa = 4.24SNVDD1081 pKa = 3.3GLYY1084 pKa = 10.19TLFKK1088 pKa = 10.78AQIAQTDD1095 pKa = 4.81LINRR1099 pKa = 11.84ITHH1102 pKa = 5.3YY1103 pKa = 10.86SVVAEE1108 pKa = 4.15YY1109 pKa = 10.84LKK1111 pKa = 10.84SLYY1114 pKa = 11.13VNDD1117 pKa = 4.78KK1118 pKa = 9.33GTLSAAGEE1126 pKa = 4.04LLNWKK1131 pKa = 8.55EE1132 pKa = 4.03WEE1134 pKa = 4.34GQQ1136 pKa = 3.33
Molecular weight: 129.36 kDa
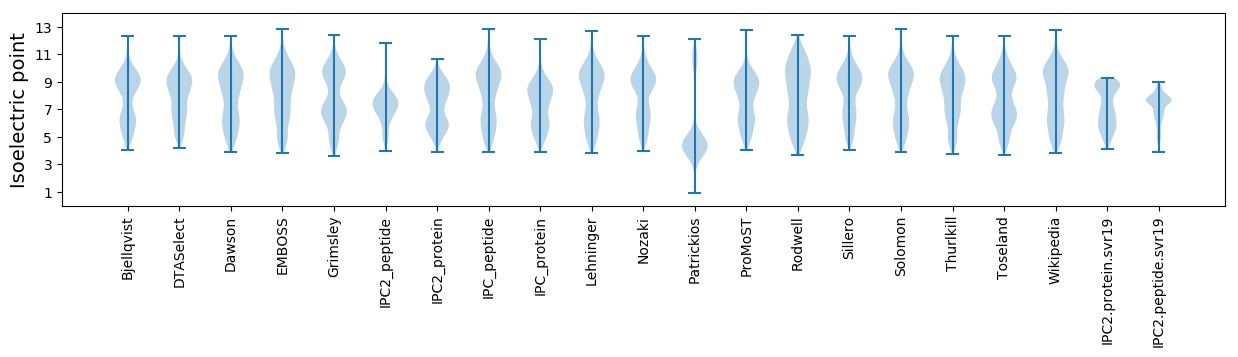
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0U3P426|A0A0U3P426_9MOLU DNA topoisomerase 1 OS=Mycoplasma sp. (ex Biomphalaria glabrata) OX=1749074 GN=topA PE=3 SV=1
MM1 pKa = 7.47LGKK4 pKa = 10.29NITNQRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84HH13 pKa = 4.6LRR15 pKa = 11.84LRR17 pKa = 11.84KK18 pKa = 9.49RR19 pKa = 11.84LIGTSNQPRR28 pKa = 11.84LCVTVSNKK36 pKa = 10.26YY37 pKa = 10.05ISAQIIDD44 pKa = 4.64DD45 pKa = 4.41SNHH48 pKa = 5.35HH49 pKa = 5.98TLAFATTQTMEE60 pKa = 4.55LANKK64 pKa = 9.94NNIEE68 pKa = 4.01AATKK72 pKa = 10.38LGEE75 pKa = 4.18VLANRR80 pKa = 11.84AKK82 pKa = 10.26EE83 pKa = 3.99KK84 pKa = 10.68NITSIIFDD92 pKa = 3.57RR93 pKa = 11.84AGRR96 pKa = 11.84KK97 pKa = 7.37YY98 pKa = 10.09HH99 pKa = 6.58GKK101 pKa = 9.03VQAFADD107 pKa = 3.6ACRR110 pKa = 11.84ANGLIFF116 pKa = 4.37
MM1 pKa = 7.47LGKK4 pKa = 10.29NITNQRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84HH13 pKa = 4.6LRR15 pKa = 11.84LRR17 pKa = 11.84KK18 pKa = 9.49RR19 pKa = 11.84LIGTSNQPRR28 pKa = 11.84LCVTVSNKK36 pKa = 10.26YY37 pKa = 10.05ISAQIIDD44 pKa = 4.64DD45 pKa = 4.41SNHH48 pKa = 5.35HH49 pKa = 5.98TLAFATTQTMEE60 pKa = 4.55LANKK64 pKa = 9.94NNIEE68 pKa = 4.01AATKK72 pKa = 10.38LGEE75 pKa = 4.18VLANRR80 pKa = 11.84AKK82 pKa = 10.26EE83 pKa = 3.99KK84 pKa = 10.68NITSIIFDD92 pKa = 3.57RR93 pKa = 11.84AGRR96 pKa = 11.84KK97 pKa = 7.37YY98 pKa = 10.09HH99 pKa = 6.58GKK101 pKa = 9.03VQAFADD107 pKa = 3.6ACRR110 pKa = 11.84ANGLIFF116 pKa = 4.37
Molecular weight: 13.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
197015 |
37 |
3066 |
355.0 |
40.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.957 ± 0.086 | 0.897 ± 0.038 |
5.24 ± 0.095 | 6.11 ± 0.12 |
5.479 ± 0.1 | 5.233 ± 0.084 |
1.772 ± 0.052 | 10.132 ± 0.14 |
8.864 ± 0.131 | 9.258 ± 0.084 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.945 ± 0.044 | 7.919 ± 0.13 |
2.911 ± 0.035 | 3.302 ± 0.064 |
2.865 ± 0.069 | 6.837 ± 0.107 |
5.572 ± 0.118 | 5.501 ± 0.08 |
1.077 ± 0.047 | 4.125 ± 0.083 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
