
Circoviridae 14 LDMD-2013
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; environmental samples
Average proteome isoelectric point is 7.31
Get precalculated fractions of proteins
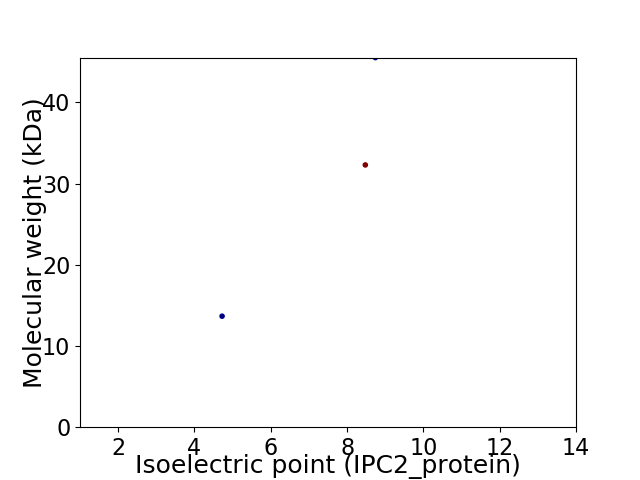
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S5TMX4|S5TMX4_9CIRC ATP-dependent helicase Rep OS=Circoviridae 14 LDMD-2013 OX=1379718 PE=3 SV=1
MM1 pKa = 7.19YY2 pKa = 10.55LVIWLEE8 pKa = 3.73WAAKK12 pKa = 9.51HH13 pKa = 5.22KK14 pKa = 10.62QKK16 pKa = 10.56EE17 pKa = 4.09LSQLTSKK24 pKa = 10.25QMNYY28 pKa = 9.83LVPGAWISEE37 pKa = 4.17LSRR40 pKa = 11.84EE41 pKa = 4.59VAWTMSSEE49 pKa = 4.63RR50 pKa = 11.84IPDD53 pKa = 3.57VFVDD57 pKa = 4.59PGPDD61 pKa = 3.67DD62 pKa = 5.64DD63 pKa = 5.33LASNTVDD70 pKa = 3.21QSNKK74 pKa = 7.38RR75 pKa = 11.84TFWDD79 pKa = 3.38WVRR82 pKa = 11.84SMDD85 pKa = 5.13SDD87 pKa = 4.58DD88 pKa = 4.46FDD90 pKa = 4.31EE91 pKa = 6.6GDD93 pKa = 3.71VKK95 pKa = 10.87KK96 pKa = 10.7LKK98 pKa = 10.42QVTEE102 pKa = 4.38SMCMPTEE109 pKa = 3.67EE110 pKa = 4.56VNRR113 pKa = 11.84HH114 pKa = 4.39NHH116 pKa = 5.22PP117 pKa = 3.62
MM1 pKa = 7.19YY2 pKa = 10.55LVIWLEE8 pKa = 3.73WAAKK12 pKa = 9.51HH13 pKa = 5.22KK14 pKa = 10.62QKK16 pKa = 10.56EE17 pKa = 4.09LSQLTSKK24 pKa = 10.25QMNYY28 pKa = 9.83LVPGAWISEE37 pKa = 4.17LSRR40 pKa = 11.84EE41 pKa = 4.59VAWTMSSEE49 pKa = 4.63RR50 pKa = 11.84IPDD53 pKa = 3.57VFVDD57 pKa = 4.59PGPDD61 pKa = 3.67DD62 pKa = 5.64DD63 pKa = 5.33LASNTVDD70 pKa = 3.21QSNKK74 pKa = 7.38RR75 pKa = 11.84TFWDD79 pKa = 3.38WVRR82 pKa = 11.84SMDD85 pKa = 5.13SDD87 pKa = 4.58DD88 pKa = 4.46FDD90 pKa = 4.31EE91 pKa = 6.6GDD93 pKa = 3.71VKK95 pKa = 10.87KK96 pKa = 10.7LKK98 pKa = 10.42QVTEE102 pKa = 4.38SMCMPTEE109 pKa = 3.67EE110 pKa = 4.56VNRR113 pKa = 11.84HH114 pKa = 4.39NHH116 pKa = 5.22PP117 pKa = 3.62
Molecular weight: 13.68 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S5TNE8|S5TNE8_9CIRC Uncharacterized protein OS=Circoviridae 14 LDMD-2013 OX=1379718 PE=4 SV=1
MM1 pKa = 7.45PRR3 pKa = 11.84YY4 pKa = 9.32GKK6 pKa = 9.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84YY10 pKa = 9.03RR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 9.89RR14 pKa = 11.84YY15 pKa = 8.13NKK17 pKa = 9.88KK18 pKa = 9.63YY19 pKa = 8.77MSRR22 pKa = 11.84FLRR25 pKa = 11.84AMLRR29 pKa = 11.84LGRR32 pKa = 11.84VRR34 pKa = 11.84TSKK37 pKa = 10.68GQSFGILTAGAGKK50 pKa = 9.91QDD52 pKa = 4.93AITVPLSARR61 pKa = 11.84IDD63 pKa = 3.83EE64 pKa = 4.91IYY66 pKa = 11.21NLLNRR71 pKa = 11.84KK72 pKa = 8.97GKK74 pKa = 10.28EE75 pKa = 3.8GEE77 pKa = 3.64IWPKK81 pKa = 10.89GFAFTIGAATVSSATHH97 pKa = 5.85ANFYY101 pKa = 10.86HH102 pKa = 7.06LGTIRR107 pKa = 11.84KK108 pKa = 7.63YY109 pKa = 8.06TFKK112 pKa = 11.22NQMNCPIEE120 pKa = 4.54AIFYY124 pKa = 9.07KK125 pKa = 10.48VQYY128 pKa = 9.83KK129 pKa = 10.44KK130 pKa = 10.7SFCSGEE136 pKa = 4.1LGSAASQAAFEE147 pKa = 4.39NFLMNAPVVLWNEE160 pKa = 4.36YY161 pKa = 9.71IGHH164 pKa = 5.3QVSTGTTASEE174 pKa = 4.23FVGVQSNAPSTVDD187 pKa = 3.27LTGDD191 pKa = 3.36YY192 pKa = 10.0DD193 pKa = 4.28AKK195 pKa = 11.4DD196 pKa = 3.9MEE198 pKa = 5.0TDD200 pKa = 3.4PQDD203 pKa = 3.59VQGEE207 pKa = 4.32DD208 pKa = 3.56VEE210 pKa = 6.33DD211 pKa = 4.36DD212 pKa = 3.93ANEE215 pKa = 4.24GGLPSGYY222 pKa = 8.76EE223 pKa = 3.41QHH225 pKa = 6.96KK226 pKa = 10.48YY227 pKa = 9.8YY228 pKa = 11.03GSGPGSSKK236 pKa = 10.65YY237 pKa = 9.48FRR239 pKa = 11.84EE240 pKa = 4.4YY241 pKa = 10.55INVIKK246 pKa = 9.78VQKK249 pKa = 10.64VVMRR253 pKa = 11.84TNQLVNISHH262 pKa = 7.4ISKK265 pKa = 10.6KK266 pKa = 7.39MRR268 pKa = 11.84KK269 pKa = 9.08YY270 pKa = 10.45DD271 pKa = 3.71AEE273 pKa = 4.05EE274 pKa = 3.86LKK276 pKa = 11.11AFANAYY282 pKa = 9.69QGASSNVGAGARR294 pKa = 11.84IHH296 pKa = 6.91RR297 pKa = 11.84KK298 pKa = 9.39GDD300 pKa = 3.11VDD302 pKa = 3.71VMVVVRR308 pKa = 11.84GLPGVRR314 pKa = 11.84TITAADD320 pKa = 3.56VTEE323 pKa = 5.17DD324 pKa = 3.15KK325 pKa = 11.34AQIGWEE331 pKa = 4.25SGVHH335 pKa = 5.05EE336 pKa = 4.56PAVATLNSNIVTTSPASLVWTHH358 pKa = 6.4QIYY361 pKa = 10.67HH362 pKa = 6.19KK363 pKa = 10.22VGWMPTGGYY372 pKa = 7.26PQHH375 pKa = 7.09SYY377 pKa = 11.18GSRR380 pKa = 11.84FYY382 pKa = 11.03YY383 pKa = 8.52PTGTYY388 pKa = 9.95EE389 pKa = 3.79VDD391 pKa = 3.14ADD393 pKa = 3.93GADD396 pKa = 3.29AHH398 pKa = 7.01LSLSTFNATKK408 pKa = 10.45VDD410 pKa = 3.73SS411 pKa = 3.97
MM1 pKa = 7.45PRR3 pKa = 11.84YY4 pKa = 9.32GKK6 pKa = 9.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84YY10 pKa = 9.03RR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 9.89RR14 pKa = 11.84YY15 pKa = 8.13NKK17 pKa = 9.88KK18 pKa = 9.63YY19 pKa = 8.77MSRR22 pKa = 11.84FLRR25 pKa = 11.84AMLRR29 pKa = 11.84LGRR32 pKa = 11.84VRR34 pKa = 11.84TSKK37 pKa = 10.68GQSFGILTAGAGKK50 pKa = 9.91QDD52 pKa = 4.93AITVPLSARR61 pKa = 11.84IDD63 pKa = 3.83EE64 pKa = 4.91IYY66 pKa = 11.21NLLNRR71 pKa = 11.84KK72 pKa = 8.97GKK74 pKa = 10.28EE75 pKa = 3.8GEE77 pKa = 3.64IWPKK81 pKa = 10.89GFAFTIGAATVSSATHH97 pKa = 5.85ANFYY101 pKa = 10.86HH102 pKa = 7.06LGTIRR107 pKa = 11.84KK108 pKa = 7.63YY109 pKa = 8.06TFKK112 pKa = 11.22NQMNCPIEE120 pKa = 4.54AIFYY124 pKa = 9.07KK125 pKa = 10.48VQYY128 pKa = 9.83KK129 pKa = 10.44KK130 pKa = 10.7SFCSGEE136 pKa = 4.1LGSAASQAAFEE147 pKa = 4.39NFLMNAPVVLWNEE160 pKa = 4.36YY161 pKa = 9.71IGHH164 pKa = 5.3QVSTGTTASEE174 pKa = 4.23FVGVQSNAPSTVDD187 pKa = 3.27LTGDD191 pKa = 3.36YY192 pKa = 10.0DD193 pKa = 4.28AKK195 pKa = 11.4DD196 pKa = 3.9MEE198 pKa = 5.0TDD200 pKa = 3.4PQDD203 pKa = 3.59VQGEE207 pKa = 4.32DD208 pKa = 3.56VEE210 pKa = 6.33DD211 pKa = 4.36DD212 pKa = 3.93ANEE215 pKa = 4.24GGLPSGYY222 pKa = 8.76EE223 pKa = 3.41QHH225 pKa = 6.96KK226 pKa = 10.48YY227 pKa = 9.8YY228 pKa = 11.03GSGPGSSKK236 pKa = 10.65YY237 pKa = 9.48FRR239 pKa = 11.84EE240 pKa = 4.4YY241 pKa = 10.55INVIKK246 pKa = 9.78VQKK249 pKa = 10.64VVMRR253 pKa = 11.84TNQLVNISHH262 pKa = 7.4ISKK265 pKa = 10.6KK266 pKa = 7.39MRR268 pKa = 11.84KK269 pKa = 9.08YY270 pKa = 10.45DD271 pKa = 3.71AEE273 pKa = 4.05EE274 pKa = 3.86LKK276 pKa = 11.11AFANAYY282 pKa = 9.69QGASSNVGAGARR294 pKa = 11.84IHH296 pKa = 6.91RR297 pKa = 11.84KK298 pKa = 9.39GDD300 pKa = 3.11VDD302 pKa = 3.71VMVVVRR308 pKa = 11.84GLPGVRR314 pKa = 11.84TITAADD320 pKa = 3.56VTEE323 pKa = 5.17DD324 pKa = 3.15KK325 pKa = 11.34AQIGWEE331 pKa = 4.25SGVHH335 pKa = 5.05EE336 pKa = 4.56PAVATLNSNIVTTSPASLVWTHH358 pKa = 6.4QIYY361 pKa = 10.67HH362 pKa = 6.19KK363 pKa = 10.22VGWMPTGGYY372 pKa = 7.26PQHH375 pKa = 7.09SYY377 pKa = 11.18GSRR380 pKa = 11.84FYY382 pKa = 11.03YY383 pKa = 8.52PTGTYY388 pKa = 9.95EE389 pKa = 3.79VDD391 pKa = 3.14ADD393 pKa = 3.93GADD396 pKa = 3.29AHH398 pKa = 7.01LSLSTFNATKK408 pKa = 10.45VDD410 pKa = 3.73SS411 pKa = 3.97
Molecular weight: 45.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
808 |
117 |
411 |
269.3 |
30.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.054 ± 1.382 | 0.619 ± 0.1 |
5.446 ± 1.352 | 5.941 ± 0.789 |
3.837 ± 0.407 | 7.302 ± 1.679 |
2.599 ± 0.051 | 5.074 ± 0.83 |
7.054 ± 0.341 | 6.683 ± 1.203 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.351 ± 0.805 | 5.322 ± 0.601 |
4.332 ± 0.467 | 3.96 ± 0.092 |
5.322 ± 0.319 | 7.054 ± 0.801 |
6.188 ± 0.355 | 6.807 ± 0.895 |
1.98 ± 0.942 | 5.074 ± 0.995 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
