
Pseudactinotalea sp. HY158
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Cellulomonadaceae; Pseudactinotalea; unclassified Pseudactinotalea
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
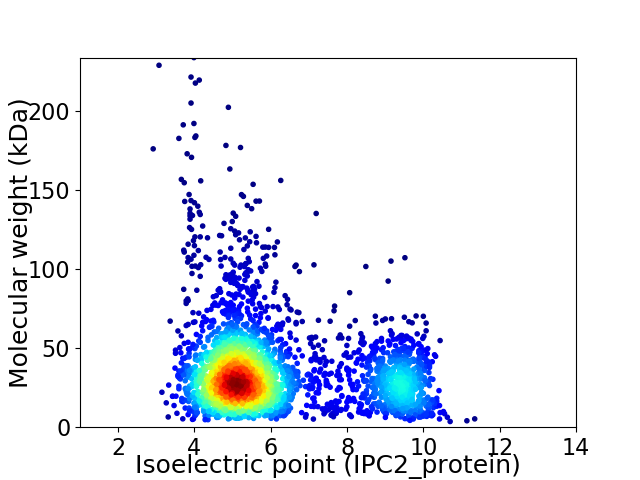
Virtual 2D-PAGE plot for 3052 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5Q2VZ47|A0A5Q2VZ47_9CELL Cytochrome bc1 complex Rieske iron-sulfur subunit OS=Pseudactinotalea sp. HY158 OX=2654547 GN=GCE65_06835 PE=4 SV=1
MM1 pKa = 7.55AATVSGCLALLTGPIAAFATTGGAGEE27 pKa = 4.44AGGNAAEE34 pKa = 4.49RR35 pKa = 11.84EE36 pKa = 4.21PARR39 pKa = 11.84LTIVSDD45 pKa = 3.2ATGAIGAGEE54 pKa = 4.53APSFSYY60 pKa = 11.13GSTSDD65 pKa = 3.42VAGSSWGEE73 pKa = 4.1GFTLAPTPEE82 pKa = 5.05DD83 pKa = 3.75PDD85 pKa = 3.87EE86 pKa = 4.31LTGALLPGEE95 pKa = 4.39AVTVEE100 pKa = 4.09AEE102 pKa = 4.28LPQGWVLDD110 pKa = 4.37DD111 pKa = 3.92LTCTGGDD118 pKa = 3.73DD119 pKa = 3.79VTVGATSVTVTSASDD134 pKa = 3.55VSCTFTNAPVATTGEE149 pKa = 4.36ADD151 pKa = 3.71PADD154 pKa = 4.4PADD157 pKa = 4.25PADD160 pKa = 4.25PADD163 pKa = 4.05PADD166 pKa = 4.39PDD168 pKa = 3.72VAEE171 pKa = 5.15PEE173 pKa = 4.36PADD176 pKa = 3.77EE177 pKa = 5.16PEE179 pKa = 4.6PADD182 pKa = 4.06APASAARR189 pKa = 11.84PSSADD194 pKa = 3.06VHH196 pKa = 6.26RR197 pKa = 11.84AGIHH201 pKa = 5.51PADD204 pKa = 4.24MQPMAVGEE212 pKa = 4.32APVCSSGYY220 pKa = 10.08VYY222 pKa = 10.71SVYY225 pKa = 10.88DD226 pKa = 3.73GGDD229 pKa = 3.25LVQIDD234 pKa = 3.9PSGAVSTMGSWSSFVDD250 pKa = 4.41GPGGSVNGLAIGDD263 pKa = 4.09GGTVVYY269 pKa = 11.1AFMRR273 pKa = 11.84FSSDD277 pKa = 2.97TAEE280 pKa = 4.85DD281 pKa = 3.89PLSNATVLRR290 pKa = 11.84YY291 pKa = 9.76LPATDD296 pKa = 3.15AWEE299 pKa = 4.22SLGTLNSNTTQQLIAGAVDD318 pKa = 4.89FASGDD323 pKa = 3.9YY324 pKa = 10.95FFGGYY329 pKa = 10.08DD330 pKa = 3.2SSNNFQLWRR339 pKa = 11.84YY340 pKa = 9.69DD341 pKa = 3.28VGTGLFDD348 pKa = 3.26KK349 pKa = 10.95VGYY352 pKa = 9.25FVSGLVGANGDD363 pKa = 3.79MAFDD367 pKa = 3.28AHH369 pKa = 6.72GNLFVIASDD378 pKa = 3.68DD379 pKa = 3.98DD380 pKa = 5.06GNVTEE385 pKa = 5.56FMITAATLAAAEE397 pKa = 4.21NNALDD402 pKa = 4.84LSQTHH407 pKa = 6.31AFDD410 pKa = 3.55VGGVAVNGVAFDD422 pKa = 4.34ADD424 pKa = 3.39GSIYY428 pKa = 10.41LGSDD432 pKa = 3.12TTVYY436 pKa = 10.53KK437 pKa = 10.64YY438 pKa = 11.01DD439 pKa = 3.47PTTWTSLGTHH449 pKa = 6.6TGNLVDD455 pKa = 3.96STDD458 pKa = 3.8LASCNSPGTITVRR471 pKa = 11.84KK472 pKa = 10.01DD473 pKa = 2.79ITLRR477 pKa = 11.84AASADD482 pKa = 3.57QFRR485 pKa = 11.84LTLEE489 pKa = 4.07RR490 pKa = 11.84DD491 pKa = 3.44AVTVATEE498 pKa = 4.26DD499 pKa = 3.42TSGTTTGLQPTQIGPVPAISGADD522 pKa = 3.44YY523 pKa = 10.93SFSEE527 pKa = 5.12AIIAFPGSSQMSAYY541 pKa = 9.15TSTWEE546 pKa = 4.21CEE548 pKa = 3.89AGGSVIATGTGTAGVVTIPPPLSGDD573 pKa = 3.21EE574 pKa = 4.23GAHH577 pKa = 4.86VVCTFTNAPKK587 pKa = 9.9TNPIEE592 pKa = 4.26FTKK595 pKa = 10.27RR596 pKa = 11.84WQGAIAGDD604 pKa = 3.77TAEE607 pKa = 4.81LSMSGFALVDD617 pKa = 3.3ATTATSTADD626 pKa = 3.39GTNPQHH632 pKa = 6.68DD633 pKa = 4.46TVNVATGAAAYY644 pKa = 9.41GSSVTLAEE652 pKa = 4.6NIGHH656 pKa = 6.72GSDD659 pKa = 3.79PAQGSYY665 pKa = 10.23NATYY669 pKa = 9.08TCTQGGVTVAVAAQTFVMPFTDD691 pKa = 3.87GTVEE695 pKa = 4.15CVVTNTPRR703 pKa = 11.84TGNIVVTKK711 pKa = 9.63KK712 pKa = 9.7WVVQDD717 pKa = 3.31SAGNVVEE724 pKa = 5.06TYY726 pKa = 10.25DD727 pKa = 3.62IPADD731 pKa = 3.78ASTLPAGLHH740 pKa = 5.84AQLTLDD746 pKa = 3.83GQNRR750 pKa = 11.84AWDD753 pKa = 3.9ASDD756 pKa = 3.02SRR758 pKa = 11.84TIGSVVTIDD767 pKa = 4.14EE768 pKa = 4.37VATLDD773 pKa = 3.69GDD775 pKa = 4.31ALPGCSILEE784 pKa = 4.09PPVLTFEE791 pKa = 4.24GTIATFAPTPGEE803 pKa = 4.34DD804 pKa = 3.0YY805 pKa = 11.17GLPFDD810 pKa = 4.08ATVAAGTSRR819 pKa = 11.84FHH821 pKa = 5.8ITNTIEE827 pKa = 4.31CTQSLSLVKK836 pKa = 10.38QVDD839 pKa = 3.86VGAVTADD846 pKa = 3.02TWTLSATKK854 pKa = 9.64PDD856 pKa = 3.15GSAALEE862 pKa = 4.79GPSGTYY868 pKa = 10.46SPTTPVTAAVTATVPYY884 pKa = 10.46ALAEE888 pKa = 4.39AGGPATYY895 pKa = 10.52VPDD898 pKa = 4.45GGWVCATSDD907 pKa = 3.82SEE909 pKa = 4.5AVAVTDD915 pKa = 3.97GAVEE919 pKa = 4.31VPLGADD925 pKa = 3.51VTCTITNTTALLTVLKK941 pKa = 10.17HH942 pKa = 6.3IDD944 pKa = 3.56GSSDD948 pKa = 3.65LSPSDD953 pKa = 3.31WDD955 pKa = 3.65LTATPDD961 pKa = 3.38AGVAGLTPTTVPGADD976 pKa = 3.78GAATDD981 pKa = 4.58PNAASTFEE989 pKa = 3.77VRR991 pKa = 11.84PGHH994 pKa = 6.21GYY996 pKa = 10.52SLAEE1000 pKa = 3.95ALAAGHH1006 pKa = 6.93EE1007 pKa = 4.28DD1008 pKa = 3.31TAYY1011 pKa = 10.87LQLALQHH1018 pKa = 6.49WDD1020 pKa = 2.95GAEE1023 pKa = 4.01WVDD1026 pKa = 3.49VDD1028 pKa = 3.99ADD1030 pKa = 4.24TVSVEE1035 pKa = 4.08AGAHH1039 pKa = 4.8DD1040 pKa = 4.08VYY1042 pKa = 11.17RR1043 pKa = 11.84FVNAGVTAISLPLTGGLGTDD1063 pKa = 4.14LFLALGALGLIAAVALAGWQHH1084 pKa = 6.27RR1085 pKa = 11.84RR1086 pKa = 11.84TTRR1089 pKa = 11.84MRR1091 pKa = 11.84RR1092 pKa = 11.84AA1093 pKa = 3.22
MM1 pKa = 7.55AATVSGCLALLTGPIAAFATTGGAGEE27 pKa = 4.44AGGNAAEE34 pKa = 4.49RR35 pKa = 11.84EE36 pKa = 4.21PARR39 pKa = 11.84LTIVSDD45 pKa = 3.2ATGAIGAGEE54 pKa = 4.53APSFSYY60 pKa = 11.13GSTSDD65 pKa = 3.42VAGSSWGEE73 pKa = 4.1GFTLAPTPEE82 pKa = 5.05DD83 pKa = 3.75PDD85 pKa = 3.87EE86 pKa = 4.31LTGALLPGEE95 pKa = 4.39AVTVEE100 pKa = 4.09AEE102 pKa = 4.28LPQGWVLDD110 pKa = 4.37DD111 pKa = 3.92LTCTGGDD118 pKa = 3.73DD119 pKa = 3.79VTVGATSVTVTSASDD134 pKa = 3.55VSCTFTNAPVATTGEE149 pKa = 4.36ADD151 pKa = 3.71PADD154 pKa = 4.4PADD157 pKa = 4.25PADD160 pKa = 4.25PADD163 pKa = 4.05PADD166 pKa = 4.39PDD168 pKa = 3.72VAEE171 pKa = 5.15PEE173 pKa = 4.36PADD176 pKa = 3.77EE177 pKa = 5.16PEE179 pKa = 4.6PADD182 pKa = 4.06APASAARR189 pKa = 11.84PSSADD194 pKa = 3.06VHH196 pKa = 6.26RR197 pKa = 11.84AGIHH201 pKa = 5.51PADD204 pKa = 4.24MQPMAVGEE212 pKa = 4.32APVCSSGYY220 pKa = 10.08VYY222 pKa = 10.71SVYY225 pKa = 10.88DD226 pKa = 3.73GGDD229 pKa = 3.25LVQIDD234 pKa = 3.9PSGAVSTMGSWSSFVDD250 pKa = 4.41GPGGSVNGLAIGDD263 pKa = 4.09GGTVVYY269 pKa = 11.1AFMRR273 pKa = 11.84FSSDD277 pKa = 2.97TAEE280 pKa = 4.85DD281 pKa = 3.89PLSNATVLRR290 pKa = 11.84YY291 pKa = 9.76LPATDD296 pKa = 3.15AWEE299 pKa = 4.22SLGTLNSNTTQQLIAGAVDD318 pKa = 4.89FASGDD323 pKa = 3.9YY324 pKa = 10.95FFGGYY329 pKa = 10.08DD330 pKa = 3.2SSNNFQLWRR339 pKa = 11.84YY340 pKa = 9.69DD341 pKa = 3.28VGTGLFDD348 pKa = 3.26KK349 pKa = 10.95VGYY352 pKa = 9.25FVSGLVGANGDD363 pKa = 3.79MAFDD367 pKa = 3.28AHH369 pKa = 6.72GNLFVIASDD378 pKa = 3.68DD379 pKa = 3.98DD380 pKa = 5.06GNVTEE385 pKa = 5.56FMITAATLAAAEE397 pKa = 4.21NNALDD402 pKa = 4.84LSQTHH407 pKa = 6.31AFDD410 pKa = 3.55VGGVAVNGVAFDD422 pKa = 4.34ADD424 pKa = 3.39GSIYY428 pKa = 10.41LGSDD432 pKa = 3.12TTVYY436 pKa = 10.53KK437 pKa = 10.64YY438 pKa = 11.01DD439 pKa = 3.47PTTWTSLGTHH449 pKa = 6.6TGNLVDD455 pKa = 3.96STDD458 pKa = 3.8LASCNSPGTITVRR471 pKa = 11.84KK472 pKa = 10.01DD473 pKa = 2.79ITLRR477 pKa = 11.84AASADD482 pKa = 3.57QFRR485 pKa = 11.84LTLEE489 pKa = 4.07RR490 pKa = 11.84DD491 pKa = 3.44AVTVATEE498 pKa = 4.26DD499 pKa = 3.42TSGTTTGLQPTQIGPVPAISGADD522 pKa = 3.44YY523 pKa = 10.93SFSEE527 pKa = 5.12AIIAFPGSSQMSAYY541 pKa = 9.15TSTWEE546 pKa = 4.21CEE548 pKa = 3.89AGGSVIATGTGTAGVVTIPPPLSGDD573 pKa = 3.21EE574 pKa = 4.23GAHH577 pKa = 4.86VVCTFTNAPKK587 pKa = 9.9TNPIEE592 pKa = 4.26FTKK595 pKa = 10.27RR596 pKa = 11.84WQGAIAGDD604 pKa = 3.77TAEE607 pKa = 4.81LSMSGFALVDD617 pKa = 3.3ATTATSTADD626 pKa = 3.39GTNPQHH632 pKa = 6.68DD633 pKa = 4.46TVNVATGAAAYY644 pKa = 9.41GSSVTLAEE652 pKa = 4.6NIGHH656 pKa = 6.72GSDD659 pKa = 3.79PAQGSYY665 pKa = 10.23NATYY669 pKa = 9.08TCTQGGVTVAVAAQTFVMPFTDD691 pKa = 3.87GTVEE695 pKa = 4.15CVVTNTPRR703 pKa = 11.84TGNIVVTKK711 pKa = 9.63KK712 pKa = 9.7WVVQDD717 pKa = 3.31SAGNVVEE724 pKa = 5.06TYY726 pKa = 10.25DD727 pKa = 3.62IPADD731 pKa = 3.78ASTLPAGLHH740 pKa = 5.84AQLTLDD746 pKa = 3.83GQNRR750 pKa = 11.84AWDD753 pKa = 3.9ASDD756 pKa = 3.02SRR758 pKa = 11.84TIGSVVTIDD767 pKa = 4.14EE768 pKa = 4.37VATLDD773 pKa = 3.69GDD775 pKa = 4.31ALPGCSILEE784 pKa = 4.09PPVLTFEE791 pKa = 4.24GTIATFAPTPGEE803 pKa = 4.34DD804 pKa = 3.0YY805 pKa = 11.17GLPFDD810 pKa = 4.08ATVAAGTSRR819 pKa = 11.84FHH821 pKa = 5.8ITNTIEE827 pKa = 4.31CTQSLSLVKK836 pKa = 10.38QVDD839 pKa = 3.86VGAVTADD846 pKa = 3.02TWTLSATKK854 pKa = 9.64PDD856 pKa = 3.15GSAALEE862 pKa = 4.79GPSGTYY868 pKa = 10.46SPTTPVTAAVTATVPYY884 pKa = 10.46ALAEE888 pKa = 4.39AGGPATYY895 pKa = 10.52VPDD898 pKa = 4.45GGWVCATSDD907 pKa = 3.82SEE909 pKa = 4.5AVAVTDD915 pKa = 3.97GAVEE919 pKa = 4.31VPLGADD925 pKa = 3.51VTCTITNTTALLTVLKK941 pKa = 10.17HH942 pKa = 6.3IDD944 pKa = 3.56GSSDD948 pKa = 3.65LSPSDD953 pKa = 3.31WDD955 pKa = 3.65LTATPDD961 pKa = 3.38AGVAGLTPTTVPGADD976 pKa = 3.78GAATDD981 pKa = 4.58PNAASTFEE989 pKa = 3.77VRR991 pKa = 11.84PGHH994 pKa = 6.21GYY996 pKa = 10.52SLAEE1000 pKa = 3.95ALAAGHH1006 pKa = 6.93EE1007 pKa = 4.28DD1008 pKa = 3.31TAYY1011 pKa = 10.87LQLALQHH1018 pKa = 6.49WDD1020 pKa = 2.95GAEE1023 pKa = 4.01WVDD1026 pKa = 3.49VDD1028 pKa = 3.99ADD1030 pKa = 4.24TVSVEE1035 pKa = 4.08AGAHH1039 pKa = 4.8DD1040 pKa = 4.08VYY1042 pKa = 11.17RR1043 pKa = 11.84FVNAGVTAISLPLTGGLGTDD1063 pKa = 4.14LFLALGALGLIAAVALAGWQHH1084 pKa = 6.27RR1085 pKa = 11.84RR1086 pKa = 11.84TTRR1089 pKa = 11.84MRR1091 pKa = 11.84RR1092 pKa = 11.84AA1093 pKa = 3.22
Molecular weight: 110.79 kDa
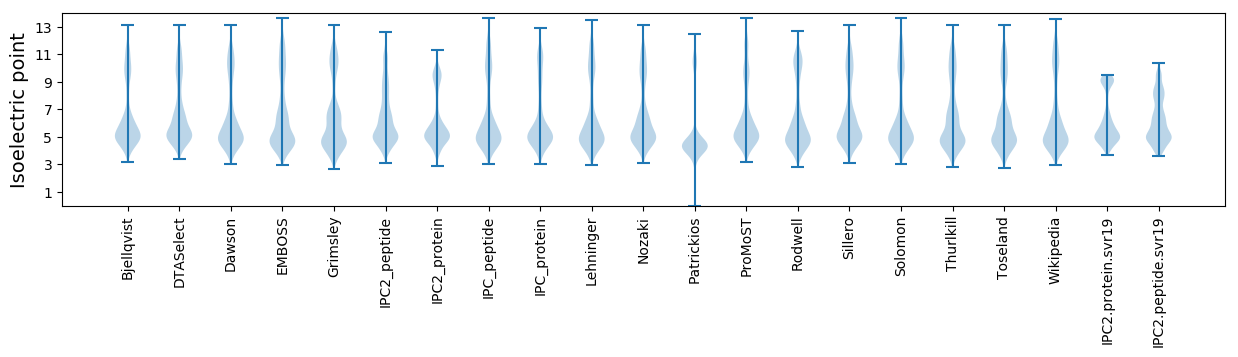
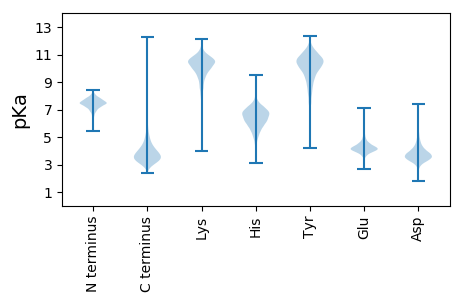
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5Q2W5T7|A0A5Q2W5T7_9CELL Oligoribonuclease OS=Pseudactinotalea sp. HY158 OX=2654547 GN=GCE65_11990 PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84ARR15 pKa = 11.84THH17 pKa = 5.69GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILSARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.77GRR40 pKa = 11.84ASVSAA45 pKa = 3.98
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84ARR15 pKa = 11.84THH17 pKa = 5.69GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILSARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.77GRR40 pKa = 11.84ASVSAA45 pKa = 3.98
Molecular weight: 5.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1062837 |
32 |
2300 |
348.2 |
37.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.99 ± 0.07 | 0.587 ± 0.012 |
6.347 ± 0.047 | 5.683 ± 0.037 |
2.735 ± 0.026 | 9.628 ± 0.047 |
2.15 ± 0.023 | 4.024 ± 0.03 |
1.44 ± 0.024 | 10.007 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.753 ± 0.018 | 1.763 ± 0.022 |
5.881 ± 0.039 | 2.559 ± 0.026 |
7.808 ± 0.06 | 5.399 ± 0.029 |
6.31 ± 0.046 | 8.532 ± 0.043 |
1.464 ± 0.018 | 1.938 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
