
Tenacibaculum sp. Bg11-29
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Tenacibaculum; unclassified Tenacibaculum
Average proteome isoelectric point is 7.15
Get precalculated fractions of proteins
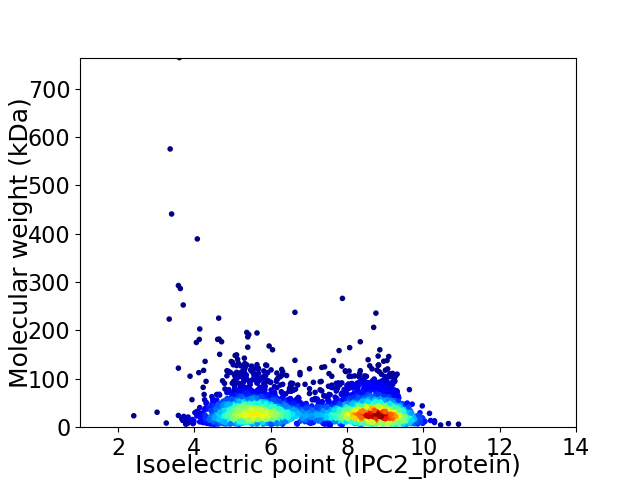
Virtual 2D-PAGE plot for 3926 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2N1FCL0|A0A2N1FCL0_9FLAO MarR family transcriptional regulator OS=Tenacibaculum sp. Bg11-29 OX=2058306 GN=CXF68_19065 PE=4 SV=1
MM1 pKa = 7.56GNLKK5 pKa = 10.41RR6 pKa = 11.84SIKK9 pKa = 10.04RR10 pKa = 11.84FKK12 pKa = 10.96LSVLFLTLSLIYY24 pKa = 10.77SNNSWAQQFYY34 pKa = 7.86PTNFDD39 pKa = 3.37VTTLNGTNGFVVPGLVTNYY58 pKa = 10.54KK59 pKa = 10.12LGRR62 pKa = 11.84EE63 pKa = 4.15VQFIGDD69 pKa = 3.61INNDD73 pKa = 3.3GFEE76 pKa = 5.24DD77 pKa = 3.42ICLGNGNQTVGVNDD91 pKa = 4.06LSGRR95 pKa = 11.84AYY97 pKa = 9.78IIFGASTGFSTPFDD111 pKa = 3.75LTSLNGTNGFIVEE124 pKa = 4.75GIGYY128 pKa = 8.57DD129 pKa = 3.55EE130 pKa = 4.66RR131 pKa = 11.84RR132 pKa = 11.84GTTVAGPGDD141 pKa = 3.84INGDD145 pKa = 3.95GIDD148 pKa = 4.28DD149 pKa = 5.35LIIGTSSSSSDD160 pKa = 3.22DD161 pKa = 3.22MVIYY165 pKa = 8.51GTTTFPALLKK175 pKa = 10.0VTDD178 pKa = 3.65INGANGFLLDD188 pKa = 4.06TPGSYY193 pKa = 9.5QVASLGDD200 pKa = 3.62VNGDD204 pKa = 4.0SINDD208 pKa = 4.24FIIGTPHH215 pKa = 7.31WSGQSWIVFGRR226 pKa = 11.84TSNYY230 pKa = 8.0PALVDD235 pKa = 3.5ATWINNAVNGFRR247 pKa = 11.84TSSFPTSRR255 pKa = 11.84PSLKK259 pKa = 10.71VGGAGDD265 pKa = 3.86INNDD269 pKa = 4.16GINDD273 pKa = 3.68ILIGGWNNSANPNSYY288 pKa = 11.42ALFGKK293 pKa = 7.95NTSFDD298 pKa = 3.43AVVNLVTVDD307 pKa = 3.44GTDD310 pKa = 3.14GFLIDD315 pKa = 4.18NEE317 pKa = 4.4GNSFITEE324 pKa = 4.05VGPLGDD330 pKa = 4.36INNDD334 pKa = 4.01GIDD337 pKa = 3.55DD338 pKa = 4.23CFSEE342 pKa = 4.21NNIIFGSTSIFPANLLMSNFNGTNGFVLEE371 pKa = 4.55NYY373 pKa = 9.51VGSAAPTGDD382 pKa = 3.5VNFDD386 pKa = 4.1GIDD389 pKa = 3.49DD390 pKa = 5.01FIVAGADD397 pKa = 3.43DD398 pKa = 4.14YY399 pKa = 12.12VVFGKK404 pKa = 8.81TGGFSPTFNSTAIDD418 pKa = 3.72GANGFKK424 pKa = 10.29IPLIGSGGGGSVDD437 pKa = 3.32GGKK440 pKa = 9.99DD441 pKa = 3.44VNGDD445 pKa = 4.05GITDD449 pKa = 4.6FIFADD454 pKa = 3.93TGAGSTGEE462 pKa = 4.21VYY464 pKa = 10.75VVFGGDD470 pKa = 3.43HH471 pKa = 6.05YY472 pKa = 12.02AMPLNATHH480 pKa = 6.65PQANNEE486 pKa = 4.18TTTGFDD492 pKa = 2.99ILVNGPEE499 pKa = 4.38TGTIHH504 pKa = 6.65YY505 pKa = 9.57VILPGTFSGAVNHH518 pKa = 5.47NTILTGTGAIANGNFLMSTANANINEE544 pKa = 4.55VISSLTAATTYY555 pKa = 10.52DD556 pKa = 3.13VHH558 pKa = 8.23LFLEE562 pKa = 4.77DD563 pKa = 4.17GIGNKK568 pKa = 10.2SIIYY572 pKa = 9.47HH573 pKa = 6.38LNNVTTLSAGDD584 pKa = 3.63ITPPSITCPANQTVTCGTTVIPDD607 pKa = 3.37YY608 pKa = 10.44TGLVTTTDD616 pKa = 3.18IVDD619 pKa = 4.15PSPVVTQSPVAGAAFVSGMIITMTSTDD646 pKa = 3.16ASLNSASCTFNVTVGGDD663 pKa = 3.56NVKK666 pKa = 8.94PTITCLISEE675 pKa = 4.46SLFINSTLPNYY686 pKa = 9.29FFKK689 pKa = 11.14LKK691 pKa = 10.83ASDD694 pKa = 3.43NCTSIFNLTYY704 pKa = 9.66TQVPAQGTVFTANTNVTVVVTDD726 pKa = 3.69EE727 pKa = 4.22NGNTEE732 pKa = 3.98TCTFLVTVKK741 pKa = 10.71GATPPVNCNTTSVNINDD758 pKa = 4.37LDD760 pKa = 4.06GDD762 pKa = 3.83NGFQIDD768 pKa = 3.77GDD770 pKa = 4.1AVKK773 pKa = 10.5AQLGHH778 pKa = 6.26DD779 pKa = 3.55VRR781 pKa = 11.84GAGDD785 pKa = 3.48VNGDD789 pKa = 4.35GIQDD793 pKa = 3.83FLVGSPNKK801 pKa = 10.25AYY803 pKa = 10.64NYY805 pKa = 7.91NAAYY809 pKa = 9.9GRR811 pKa = 11.84EE812 pKa = 3.91RR813 pKa = 11.84GNFPGNVFVVFGKK826 pKa = 10.93GNNFLPNIDD835 pKa = 3.82TVLLDD840 pKa = 3.53GTDD843 pKa = 3.15GFKK846 pKa = 9.99IHH848 pKa = 6.87NDD850 pKa = 2.88IPNGGSQYY858 pKa = 11.1SGYY861 pKa = 9.97QVSTAGDD868 pKa = 3.58INNDD872 pKa = 4.12GIDD875 pKa = 4.17DD876 pKa = 4.71LMFSDD881 pKa = 4.45PFRR884 pKa = 11.84RR885 pKa = 11.84NPLGSEE891 pKa = 4.02LGTIYY896 pKa = 11.03VVFGKK901 pKa = 7.45TTGFLPVFNLSNINGTNGFVFLGTDD926 pKa = 3.94AYY928 pKa = 10.33DD929 pKa = 3.52QSGKK933 pKa = 10.85SIDD936 pKa = 3.7SAGDD940 pKa = 3.39INNDD944 pKa = 4.02GINDD948 pKa = 4.4IIIGASTGRR957 pKa = 11.84GTNNSGTCYY966 pKa = 10.4VIYY969 pKa = 10.08GKK971 pKa = 8.7TGSFPALVRR980 pKa = 11.84GSDD983 pKa = 3.73LNGTNGFIIKK993 pKa = 10.23GDD995 pKa = 3.57TDD997 pKa = 4.07GEE999 pKa = 4.3QIGYY1003 pKa = 9.83GVVGLGDD1010 pKa = 3.68VNGDD1014 pKa = 3.95GIDD1017 pKa = 3.64DD1018 pKa = 4.41LGFEE1022 pKa = 4.63GGGKK1026 pKa = 9.95KK1027 pKa = 8.25KK1028 pKa = 9.39TYY1030 pKa = 10.92VLFGKK1035 pKa = 8.55TGSFPAIVATATINGTNGFYY1055 pKa = 11.08ADD1057 pKa = 3.44ISSFTSSYY1065 pKa = 9.31YY1066 pKa = 10.4RR1067 pKa = 11.84SITGVGDD1074 pKa = 3.44VNNDD1078 pKa = 3.85GIKK1081 pKa = 11.04DD1082 pKa = 3.51MLFGNRR1088 pKa = 11.84FLLFGRR1094 pKa = 11.84NTGFTTAEE1102 pKa = 5.04DD1103 pKa = 3.39ITNLDD1108 pKa = 3.86GIKK1111 pKa = 10.09GVEE1114 pKa = 4.88FIGSTYY1120 pKa = 10.29RR1121 pKa = 11.84KK1122 pKa = 9.52SSAGGDD1128 pKa = 3.8FNNDD1132 pKa = 3.72GIDD1135 pKa = 3.5DD1136 pKa = 5.18LIIGRR1141 pKa = 11.84SSSSIAIVYY1150 pKa = 8.36GKK1152 pKa = 8.36STPWDD1157 pKa = 3.24ATLNPFSLPSSEE1169 pKa = 4.05AFKK1172 pKa = 10.68VTGLSSDD1179 pKa = 3.75YY1180 pKa = 11.02SVSDD1184 pKa = 3.61LADD1187 pKa = 3.3INGDD1191 pKa = 3.71GITDD1195 pKa = 3.81VIVGEE1200 pKa = 4.27KK1201 pKa = 10.02EE1202 pKa = 3.44RR1203 pKa = 11.84LYY1205 pKa = 10.93YY1206 pKa = 10.57DD1207 pKa = 3.44SSTSLNPGRR1216 pKa = 11.84ANVIYY1221 pKa = 9.54GFSVIDD1227 pKa = 3.48TEE1229 pKa = 5.19APVITCPIDD1238 pKa = 3.29QVLVSGSLLPDD1249 pKa = 4.23YY1250 pKa = 10.79ISLATVTDD1258 pKa = 3.62NCDD1261 pKa = 3.2TSPVITQAPVAGGVYY1276 pKa = 9.17TPGMTITLTATDD1288 pKa = 3.95KK1289 pKa = 11.12KK1290 pKa = 11.0GNSANCDD1297 pKa = 4.08FIVTDD1302 pKa = 3.62NTDD1305 pKa = 3.76TIGPTATNPLPINVQCLDD1323 pKa = 4.51DD1324 pKa = 4.4VPLPNPLVVTDD1335 pKa = 3.95EE1336 pKa = 4.78ADD1338 pKa = 3.22NSGIPPTVAFVSDD1351 pKa = 4.15VNTSPSVITRR1361 pKa = 11.84TYY1363 pKa = 10.91SVTDD1367 pKa = 3.62GAGNTINVTQTITISDD1383 pKa = 4.38TIKK1386 pKa = 9.61PTATNPLPINVSCIGDD1402 pKa = 3.77VPTADD1407 pKa = 2.77ITVVTDD1413 pKa = 3.4EE1414 pKa = 5.24ADD1416 pKa = 3.43NCTGTITIAFVSDD1429 pKa = 3.73VNTSPGLITRR1439 pKa = 11.84TYY1441 pKa = 10.87SVTDD1445 pKa = 3.38VAGNTVDD1452 pKa = 3.29VTQTINISDD1461 pKa = 4.38TIKK1464 pKa = 9.66PTATNPLPINISCIGDD1480 pKa = 3.64VPTADD1485 pKa = 2.8ITVVSDD1491 pKa = 4.8EE1492 pKa = 4.71IDD1494 pKa = 3.42NCTGTITVAFVSDD1507 pKa = 4.16VNTSPSVITRR1517 pKa = 11.84TYY1519 pKa = 10.86SVTDD1523 pKa = 3.33VAGNTINVTQIINVSDD1539 pKa = 3.42IVKK1542 pKa = 8.72PTASDD1547 pKa = 3.71PLPISISCIGDD1558 pKa = 3.26VPAADD1563 pKa = 3.04ITVVTDD1569 pKa = 4.66EE1570 pKa = 5.07IDD1572 pKa = 3.37NCTGTIIVAFVSDD1585 pKa = 4.19VNTSPGLITRR1595 pKa = 11.84TYY1597 pKa = 10.79SVTDD1601 pKa = 3.42AAGNTINVTQTINVSDD1617 pKa = 3.67IVKK1620 pKa = 8.72PTASDD1625 pKa = 3.65PLPINVSCAGDD1636 pKa = 3.66IPTADD1641 pKa = 2.95ITVVTDD1647 pKa = 3.4EE1648 pKa = 5.24ADD1650 pKa = 3.39NCTGTIIVAFVSDD1663 pKa = 4.22VNTSPGIITRR1673 pKa = 11.84TYY1675 pKa = 10.88SVTDD1679 pKa = 3.42AAGNTINVTQTINVSDD1695 pKa = 3.67IVKK1698 pKa = 8.72PTASDD1703 pKa = 3.65PLPINVSCAGDD1714 pKa = 3.56IPAADD1719 pKa = 3.2ITVVIDD1725 pKa = 3.78EE1726 pKa = 5.24ADD1728 pKa = 3.49NCTGAIIVAFVSDD1741 pKa = 4.22VNTSPGIITRR1751 pKa = 11.84TYY1753 pKa = 10.88SVTDD1757 pKa = 3.45AAGNTIDD1764 pKa = 3.44VTQTINVSDD1773 pKa = 3.67IVKK1776 pKa = 8.72PTASDD1781 pKa = 3.67PLPINISCIGDD1792 pKa = 3.64VPTADD1797 pKa = 2.77ITVVTDD1803 pKa = 3.4EE1804 pKa = 5.19ADD1806 pKa = 3.48NCTGAIMVAFVSDD1819 pKa = 4.05VNTSSSIITRR1829 pKa = 11.84TYY1831 pKa = 10.93SVTDD1835 pKa = 3.42AAGNTINVTQTINVSDD1851 pKa = 4.46IIKK1854 pKa = 8.71PTASDD1859 pKa = 3.92PLPINISCIGDD1870 pKa = 3.87LPVADD1875 pKa = 3.52ITVVTDD1881 pKa = 3.4EE1882 pKa = 5.19ADD1884 pKa = 3.43NCTGAITVAFVSDD1897 pKa = 4.04VNTSSSVITRR1907 pKa = 11.84TYY1909 pKa = 10.89SVTDD1913 pKa = 3.3VAGNSINVTQTINVSDD1929 pKa = 4.46IIKK1932 pKa = 8.71PTASDD1937 pKa = 3.92PLPINISCIGDD1948 pKa = 3.59VPVADD1953 pKa = 3.32ITVVTDD1959 pKa = 3.74AADD1962 pKa = 3.42NCTGVITVAFVSDD1975 pKa = 4.16VNTSPSVIIRR1985 pKa = 11.84TYY1987 pKa = 11.0SVTDD1991 pKa = 3.31VAGNSINVTQTITVSDD2007 pKa = 4.69TISPTATNPLPINVLCTGDD2026 pKa = 4.39IPTADD2031 pKa = 3.02ISAITDD2037 pKa = 3.36EE2038 pKa = 5.09ADD2040 pKa = 3.0NCTGVITVAFVNDD2053 pKa = 3.91MNTSPGVITRR2063 pKa = 11.84TYY2065 pKa = 10.86SVTDD2069 pKa = 3.39AAGNSTDD2076 pKa = 3.28VTQTITVSDD2085 pKa = 5.02IIPPTATNPAAIIISCVGDD2104 pKa = 3.86IPAADD2109 pKa = 3.33ITVVAGEE2116 pKa = 4.24VDD2118 pKa = 3.5NCTGTITVAFVSDD2131 pKa = 3.77VSDD2134 pKa = 3.89GNSNPEE2140 pKa = 4.16IITRR2144 pKa = 11.84TYY2146 pKa = 11.09SVTDD2150 pKa = 3.35AVGNFTNVTQTIIVNDD2166 pKa = 3.55STAPITTCPSNSTQKK2181 pKa = 10.27VDD2183 pKa = 3.42SGLLTAVVTFTNPVATDD2200 pKa = 3.17NCTVATINQTAGLPSGSEE2218 pKa = 3.91FPIGVNTISFEE2229 pKa = 4.19ATDD2232 pKa = 3.63AAGNTATCSFTITIEE2247 pKa = 4.22AEE2249 pKa = 4.07DD2250 pKa = 4.35PLVCTVDD2257 pKa = 2.98AGTDD2261 pKa = 3.69FDD2263 pKa = 5.08IIEE2266 pKa = 4.42GDD2268 pKa = 3.75EE2269 pKa = 4.16VALNAVISGAGVIEE2283 pKa = 4.3WSPSVGVSSTTIANPIASPTEE2304 pKa = 3.87TTTYY2308 pKa = 10.01TIRR2311 pKa = 11.84FVNAEE2316 pKa = 3.92GCVAEE2321 pKa = 4.2DD2322 pKa = 4.07TITVFVTPLEE2332 pKa = 4.19KK2333 pKa = 10.94DD2334 pKa = 3.03EE2335 pKa = 4.37TKK2337 pKa = 11.01YY2338 pKa = 11.1GFSPDD2343 pKa = 3.05GDD2345 pKa = 4.23GINEE2349 pKa = 3.8YY2350 pKa = 10.29WEE2352 pKa = 3.84IDD2354 pKa = 3.87GIEE2357 pKa = 4.31NYY2359 pKa = 10.01PNNQVLIYY2367 pKa = 10.32NRR2369 pKa = 11.84WGDD2372 pKa = 4.47LIFEE2376 pKa = 4.39TTGYY2380 pKa = 10.9NNASNVFRR2388 pKa = 11.84GIANKK2393 pKa = 9.61KK2394 pKa = 9.23RR2395 pKa = 11.84NLGAGKK2401 pKa = 10.03LPEE2404 pKa = 4.25GTYY2407 pKa = 10.65LFHH2410 pKa = 8.07IKK2412 pKa = 10.12IKK2414 pKa = 10.54GSHH2417 pKa = 5.32NLKK2420 pKa = 10.12KK2421 pKa = 10.79EE2422 pKa = 3.73KK2423 pKa = 10.68GFLILKK2429 pKa = 9.86RR2430 pKa = 3.77
MM1 pKa = 7.56GNLKK5 pKa = 10.41RR6 pKa = 11.84SIKK9 pKa = 10.04RR10 pKa = 11.84FKK12 pKa = 10.96LSVLFLTLSLIYY24 pKa = 10.77SNNSWAQQFYY34 pKa = 7.86PTNFDD39 pKa = 3.37VTTLNGTNGFVVPGLVTNYY58 pKa = 10.54KK59 pKa = 10.12LGRR62 pKa = 11.84EE63 pKa = 4.15VQFIGDD69 pKa = 3.61INNDD73 pKa = 3.3GFEE76 pKa = 5.24DD77 pKa = 3.42ICLGNGNQTVGVNDD91 pKa = 4.06LSGRR95 pKa = 11.84AYY97 pKa = 9.78IIFGASTGFSTPFDD111 pKa = 3.75LTSLNGTNGFIVEE124 pKa = 4.75GIGYY128 pKa = 8.57DD129 pKa = 3.55EE130 pKa = 4.66RR131 pKa = 11.84RR132 pKa = 11.84GTTVAGPGDD141 pKa = 3.84INGDD145 pKa = 3.95GIDD148 pKa = 4.28DD149 pKa = 5.35LIIGTSSSSSDD160 pKa = 3.22DD161 pKa = 3.22MVIYY165 pKa = 8.51GTTTFPALLKK175 pKa = 10.0VTDD178 pKa = 3.65INGANGFLLDD188 pKa = 4.06TPGSYY193 pKa = 9.5QVASLGDD200 pKa = 3.62VNGDD204 pKa = 4.0SINDD208 pKa = 4.24FIIGTPHH215 pKa = 7.31WSGQSWIVFGRR226 pKa = 11.84TSNYY230 pKa = 8.0PALVDD235 pKa = 3.5ATWINNAVNGFRR247 pKa = 11.84TSSFPTSRR255 pKa = 11.84PSLKK259 pKa = 10.71VGGAGDD265 pKa = 3.86INNDD269 pKa = 4.16GINDD273 pKa = 3.68ILIGGWNNSANPNSYY288 pKa = 11.42ALFGKK293 pKa = 7.95NTSFDD298 pKa = 3.43AVVNLVTVDD307 pKa = 3.44GTDD310 pKa = 3.14GFLIDD315 pKa = 4.18NEE317 pKa = 4.4GNSFITEE324 pKa = 4.05VGPLGDD330 pKa = 4.36INNDD334 pKa = 4.01GIDD337 pKa = 3.55DD338 pKa = 4.23CFSEE342 pKa = 4.21NNIIFGSTSIFPANLLMSNFNGTNGFVLEE371 pKa = 4.55NYY373 pKa = 9.51VGSAAPTGDD382 pKa = 3.5VNFDD386 pKa = 4.1GIDD389 pKa = 3.49DD390 pKa = 5.01FIVAGADD397 pKa = 3.43DD398 pKa = 4.14YY399 pKa = 12.12VVFGKK404 pKa = 8.81TGGFSPTFNSTAIDD418 pKa = 3.72GANGFKK424 pKa = 10.29IPLIGSGGGGSVDD437 pKa = 3.32GGKK440 pKa = 9.99DD441 pKa = 3.44VNGDD445 pKa = 4.05GITDD449 pKa = 4.6FIFADD454 pKa = 3.93TGAGSTGEE462 pKa = 4.21VYY464 pKa = 10.75VVFGGDD470 pKa = 3.43HH471 pKa = 6.05YY472 pKa = 12.02AMPLNATHH480 pKa = 6.65PQANNEE486 pKa = 4.18TTTGFDD492 pKa = 2.99ILVNGPEE499 pKa = 4.38TGTIHH504 pKa = 6.65YY505 pKa = 9.57VILPGTFSGAVNHH518 pKa = 5.47NTILTGTGAIANGNFLMSTANANINEE544 pKa = 4.55VISSLTAATTYY555 pKa = 10.52DD556 pKa = 3.13VHH558 pKa = 8.23LFLEE562 pKa = 4.77DD563 pKa = 4.17GIGNKK568 pKa = 10.2SIIYY572 pKa = 9.47HH573 pKa = 6.38LNNVTTLSAGDD584 pKa = 3.63ITPPSITCPANQTVTCGTTVIPDD607 pKa = 3.37YY608 pKa = 10.44TGLVTTTDD616 pKa = 3.18IVDD619 pKa = 4.15PSPVVTQSPVAGAAFVSGMIITMTSTDD646 pKa = 3.16ASLNSASCTFNVTVGGDD663 pKa = 3.56NVKK666 pKa = 8.94PTITCLISEE675 pKa = 4.46SLFINSTLPNYY686 pKa = 9.29FFKK689 pKa = 11.14LKK691 pKa = 10.83ASDD694 pKa = 3.43NCTSIFNLTYY704 pKa = 9.66TQVPAQGTVFTANTNVTVVVTDD726 pKa = 3.69EE727 pKa = 4.22NGNTEE732 pKa = 3.98TCTFLVTVKK741 pKa = 10.71GATPPVNCNTTSVNINDD758 pKa = 4.37LDD760 pKa = 4.06GDD762 pKa = 3.83NGFQIDD768 pKa = 3.77GDD770 pKa = 4.1AVKK773 pKa = 10.5AQLGHH778 pKa = 6.26DD779 pKa = 3.55VRR781 pKa = 11.84GAGDD785 pKa = 3.48VNGDD789 pKa = 4.35GIQDD793 pKa = 3.83FLVGSPNKK801 pKa = 10.25AYY803 pKa = 10.64NYY805 pKa = 7.91NAAYY809 pKa = 9.9GRR811 pKa = 11.84EE812 pKa = 3.91RR813 pKa = 11.84GNFPGNVFVVFGKK826 pKa = 10.93GNNFLPNIDD835 pKa = 3.82TVLLDD840 pKa = 3.53GTDD843 pKa = 3.15GFKK846 pKa = 9.99IHH848 pKa = 6.87NDD850 pKa = 2.88IPNGGSQYY858 pKa = 11.1SGYY861 pKa = 9.97QVSTAGDD868 pKa = 3.58INNDD872 pKa = 4.12GIDD875 pKa = 4.17DD876 pKa = 4.71LMFSDD881 pKa = 4.45PFRR884 pKa = 11.84RR885 pKa = 11.84NPLGSEE891 pKa = 4.02LGTIYY896 pKa = 11.03VVFGKK901 pKa = 7.45TTGFLPVFNLSNINGTNGFVFLGTDD926 pKa = 3.94AYY928 pKa = 10.33DD929 pKa = 3.52QSGKK933 pKa = 10.85SIDD936 pKa = 3.7SAGDD940 pKa = 3.39INNDD944 pKa = 4.02GINDD948 pKa = 4.4IIIGASTGRR957 pKa = 11.84GTNNSGTCYY966 pKa = 10.4VIYY969 pKa = 10.08GKK971 pKa = 8.7TGSFPALVRR980 pKa = 11.84GSDD983 pKa = 3.73LNGTNGFIIKK993 pKa = 10.23GDD995 pKa = 3.57TDD997 pKa = 4.07GEE999 pKa = 4.3QIGYY1003 pKa = 9.83GVVGLGDD1010 pKa = 3.68VNGDD1014 pKa = 3.95GIDD1017 pKa = 3.64DD1018 pKa = 4.41LGFEE1022 pKa = 4.63GGGKK1026 pKa = 9.95KK1027 pKa = 8.25KK1028 pKa = 9.39TYY1030 pKa = 10.92VLFGKK1035 pKa = 8.55TGSFPAIVATATINGTNGFYY1055 pKa = 11.08ADD1057 pKa = 3.44ISSFTSSYY1065 pKa = 9.31YY1066 pKa = 10.4RR1067 pKa = 11.84SITGVGDD1074 pKa = 3.44VNNDD1078 pKa = 3.85GIKK1081 pKa = 11.04DD1082 pKa = 3.51MLFGNRR1088 pKa = 11.84FLLFGRR1094 pKa = 11.84NTGFTTAEE1102 pKa = 5.04DD1103 pKa = 3.39ITNLDD1108 pKa = 3.86GIKK1111 pKa = 10.09GVEE1114 pKa = 4.88FIGSTYY1120 pKa = 10.29RR1121 pKa = 11.84KK1122 pKa = 9.52SSAGGDD1128 pKa = 3.8FNNDD1132 pKa = 3.72GIDD1135 pKa = 3.5DD1136 pKa = 5.18LIIGRR1141 pKa = 11.84SSSSIAIVYY1150 pKa = 8.36GKK1152 pKa = 8.36STPWDD1157 pKa = 3.24ATLNPFSLPSSEE1169 pKa = 4.05AFKK1172 pKa = 10.68VTGLSSDD1179 pKa = 3.75YY1180 pKa = 11.02SVSDD1184 pKa = 3.61LADD1187 pKa = 3.3INGDD1191 pKa = 3.71GITDD1195 pKa = 3.81VIVGEE1200 pKa = 4.27KK1201 pKa = 10.02EE1202 pKa = 3.44RR1203 pKa = 11.84LYY1205 pKa = 10.93YY1206 pKa = 10.57DD1207 pKa = 3.44SSTSLNPGRR1216 pKa = 11.84ANVIYY1221 pKa = 9.54GFSVIDD1227 pKa = 3.48TEE1229 pKa = 5.19APVITCPIDD1238 pKa = 3.29QVLVSGSLLPDD1249 pKa = 4.23YY1250 pKa = 10.79ISLATVTDD1258 pKa = 3.62NCDD1261 pKa = 3.2TSPVITQAPVAGGVYY1276 pKa = 9.17TPGMTITLTATDD1288 pKa = 3.95KK1289 pKa = 11.12KK1290 pKa = 11.0GNSANCDD1297 pKa = 4.08FIVTDD1302 pKa = 3.62NTDD1305 pKa = 3.76TIGPTATNPLPINVQCLDD1323 pKa = 4.51DD1324 pKa = 4.4VPLPNPLVVTDD1335 pKa = 3.95EE1336 pKa = 4.78ADD1338 pKa = 3.22NSGIPPTVAFVSDD1351 pKa = 4.15VNTSPSVITRR1361 pKa = 11.84TYY1363 pKa = 10.91SVTDD1367 pKa = 3.62GAGNTINVTQTITISDD1383 pKa = 4.38TIKK1386 pKa = 9.61PTATNPLPINVSCIGDD1402 pKa = 3.77VPTADD1407 pKa = 2.77ITVVTDD1413 pKa = 3.4EE1414 pKa = 5.24ADD1416 pKa = 3.43NCTGTITIAFVSDD1429 pKa = 3.73VNTSPGLITRR1439 pKa = 11.84TYY1441 pKa = 10.87SVTDD1445 pKa = 3.38VAGNTVDD1452 pKa = 3.29VTQTINISDD1461 pKa = 4.38TIKK1464 pKa = 9.66PTATNPLPINISCIGDD1480 pKa = 3.64VPTADD1485 pKa = 2.8ITVVSDD1491 pKa = 4.8EE1492 pKa = 4.71IDD1494 pKa = 3.42NCTGTITVAFVSDD1507 pKa = 4.16VNTSPSVITRR1517 pKa = 11.84TYY1519 pKa = 10.86SVTDD1523 pKa = 3.33VAGNTINVTQIINVSDD1539 pKa = 3.42IVKK1542 pKa = 8.72PTASDD1547 pKa = 3.71PLPISISCIGDD1558 pKa = 3.26VPAADD1563 pKa = 3.04ITVVTDD1569 pKa = 4.66EE1570 pKa = 5.07IDD1572 pKa = 3.37NCTGTIIVAFVSDD1585 pKa = 4.19VNTSPGLITRR1595 pKa = 11.84TYY1597 pKa = 10.79SVTDD1601 pKa = 3.42AAGNTINVTQTINVSDD1617 pKa = 3.67IVKK1620 pKa = 8.72PTASDD1625 pKa = 3.65PLPINVSCAGDD1636 pKa = 3.66IPTADD1641 pKa = 2.95ITVVTDD1647 pKa = 3.4EE1648 pKa = 5.24ADD1650 pKa = 3.39NCTGTIIVAFVSDD1663 pKa = 4.22VNTSPGIITRR1673 pKa = 11.84TYY1675 pKa = 10.88SVTDD1679 pKa = 3.42AAGNTINVTQTINVSDD1695 pKa = 3.67IVKK1698 pKa = 8.72PTASDD1703 pKa = 3.65PLPINVSCAGDD1714 pKa = 3.56IPAADD1719 pKa = 3.2ITVVIDD1725 pKa = 3.78EE1726 pKa = 5.24ADD1728 pKa = 3.49NCTGAIIVAFVSDD1741 pKa = 4.22VNTSPGIITRR1751 pKa = 11.84TYY1753 pKa = 10.88SVTDD1757 pKa = 3.45AAGNTIDD1764 pKa = 3.44VTQTINVSDD1773 pKa = 3.67IVKK1776 pKa = 8.72PTASDD1781 pKa = 3.67PLPINISCIGDD1792 pKa = 3.64VPTADD1797 pKa = 2.77ITVVTDD1803 pKa = 3.4EE1804 pKa = 5.19ADD1806 pKa = 3.48NCTGAIMVAFVSDD1819 pKa = 4.05VNTSSSIITRR1829 pKa = 11.84TYY1831 pKa = 10.93SVTDD1835 pKa = 3.42AAGNTINVTQTINVSDD1851 pKa = 4.46IIKK1854 pKa = 8.71PTASDD1859 pKa = 3.92PLPINISCIGDD1870 pKa = 3.87LPVADD1875 pKa = 3.52ITVVTDD1881 pKa = 3.4EE1882 pKa = 5.19ADD1884 pKa = 3.43NCTGAITVAFVSDD1897 pKa = 4.04VNTSSSVITRR1907 pKa = 11.84TYY1909 pKa = 10.89SVTDD1913 pKa = 3.3VAGNSINVTQTINVSDD1929 pKa = 4.46IIKK1932 pKa = 8.71PTASDD1937 pKa = 3.92PLPINISCIGDD1948 pKa = 3.59VPVADD1953 pKa = 3.32ITVVTDD1959 pKa = 3.74AADD1962 pKa = 3.42NCTGVITVAFVSDD1975 pKa = 4.16VNTSPSVIIRR1985 pKa = 11.84TYY1987 pKa = 11.0SVTDD1991 pKa = 3.31VAGNSINVTQTITVSDD2007 pKa = 4.69TISPTATNPLPINVLCTGDD2026 pKa = 4.39IPTADD2031 pKa = 3.02ISAITDD2037 pKa = 3.36EE2038 pKa = 5.09ADD2040 pKa = 3.0NCTGVITVAFVNDD2053 pKa = 3.91MNTSPGVITRR2063 pKa = 11.84TYY2065 pKa = 10.86SVTDD2069 pKa = 3.39AAGNSTDD2076 pKa = 3.28VTQTITVSDD2085 pKa = 5.02IIPPTATNPAAIIISCVGDD2104 pKa = 3.86IPAADD2109 pKa = 3.33ITVVAGEE2116 pKa = 4.24VDD2118 pKa = 3.5NCTGTITVAFVSDD2131 pKa = 3.77VSDD2134 pKa = 3.89GNSNPEE2140 pKa = 4.16IITRR2144 pKa = 11.84TYY2146 pKa = 11.09SVTDD2150 pKa = 3.35AVGNFTNVTQTIIVNDD2166 pKa = 3.55STAPITTCPSNSTQKK2181 pKa = 10.27VDD2183 pKa = 3.42SGLLTAVVTFTNPVATDD2200 pKa = 3.17NCTVATINQTAGLPSGSEE2218 pKa = 3.91FPIGVNTISFEE2229 pKa = 4.19ATDD2232 pKa = 3.63AAGNTATCSFTITIEE2247 pKa = 4.22AEE2249 pKa = 4.07DD2250 pKa = 4.35PLVCTVDD2257 pKa = 2.98AGTDD2261 pKa = 3.69FDD2263 pKa = 5.08IIEE2266 pKa = 4.42GDD2268 pKa = 3.75EE2269 pKa = 4.16VALNAVISGAGVIEE2283 pKa = 4.3WSPSVGVSSTTIANPIASPTEE2304 pKa = 3.87TTTYY2308 pKa = 10.01TIRR2311 pKa = 11.84FVNAEE2316 pKa = 3.92GCVAEE2321 pKa = 4.2DD2322 pKa = 4.07TITVFVTPLEE2332 pKa = 4.19KK2333 pKa = 10.94DD2334 pKa = 3.03EE2335 pKa = 4.37TKK2337 pKa = 11.01YY2338 pKa = 11.1GFSPDD2343 pKa = 3.05GDD2345 pKa = 4.23GINEE2349 pKa = 3.8YY2350 pKa = 10.29WEE2352 pKa = 3.84IDD2354 pKa = 3.87GIEE2357 pKa = 4.31NYY2359 pKa = 10.01PNNQVLIYY2367 pKa = 10.32NRR2369 pKa = 11.84WGDD2372 pKa = 4.47LIFEE2376 pKa = 4.39TTGYY2380 pKa = 10.9NNASNVFRR2388 pKa = 11.84GIANKK2393 pKa = 9.61KK2394 pKa = 9.23RR2395 pKa = 11.84NLGAGKK2401 pKa = 10.03LPEE2404 pKa = 4.25GTYY2407 pKa = 10.65LFHH2410 pKa = 8.07IKK2412 pKa = 10.12IKK2414 pKa = 10.54GSHH2417 pKa = 5.32NLKK2420 pKa = 10.12KK2421 pKa = 10.79EE2422 pKa = 3.73KK2423 pKa = 10.68GFLILKK2429 pKa = 9.86RR2430 pKa = 3.77
Molecular weight: 252.69 kDa
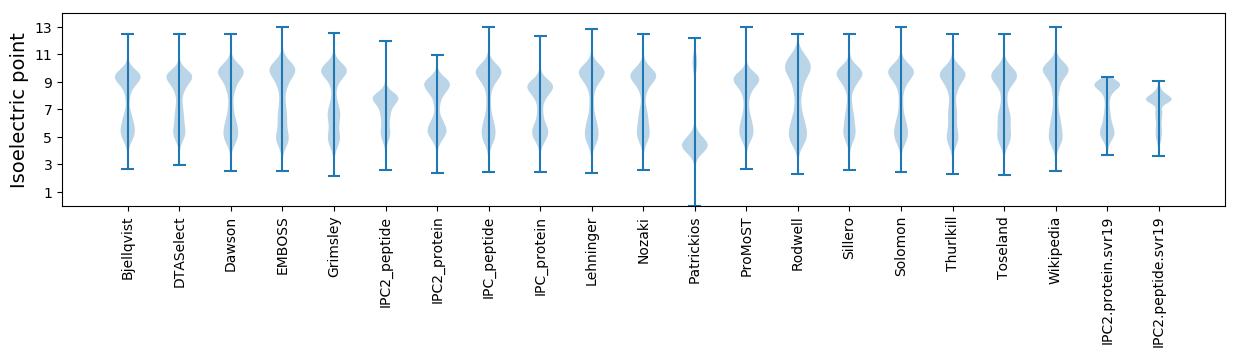
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2N1FBU1|A0A2N1FBU1_9FLAO 3-isopropylmalate dehydratase large subunit OS=Tenacibaculum sp. Bg11-29 OX=2058306 GN=leuC PE=3 SV=1
MM1 pKa = 8.03PKK3 pKa = 8.97RR4 pKa = 11.84TYY6 pKa = 10.33QPSKK10 pKa = 9.01RR11 pKa = 11.84KK12 pKa = 9.47RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.03GRR40 pKa = 11.84KK41 pKa = 7.95NLSVSSAPRR50 pKa = 11.84PKK52 pKa = 10.38KK53 pKa = 10.5
MM1 pKa = 8.03PKK3 pKa = 8.97RR4 pKa = 11.84TYY6 pKa = 10.33QPSKK10 pKa = 9.01RR11 pKa = 11.84KK12 pKa = 9.47RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.03GRR40 pKa = 11.84KK41 pKa = 7.95NLSVSSAPRR50 pKa = 11.84PKK52 pKa = 10.38KK53 pKa = 10.5
Molecular weight: 6.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1305142 |
35 |
7359 |
332.4 |
37.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.707 ± 0.042 | 0.783 ± 0.019 |
5.174 ± 0.042 | 6.397 ± 0.048 |
5.301 ± 0.04 | 6.001 ± 0.049 |
1.652 ± 0.02 | 8.623 ± 0.044 |
9.109 ± 0.091 | 9.182 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.931 ± 0.022 | 6.946 ± 0.04 |
3.043 ± 0.025 | 3.07 ± 0.022 |
3.029 ± 0.025 | 6.709 ± 0.038 |
6.212 ± 0.087 | 6.004 ± 0.046 |
1.001 ± 0.014 | 4.126 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
