
Mesorhizobium sp. 8
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Phyllobacteriaceae; Mesorhizobium; unclassified Mesorhizobium
Average proteome isoelectric point is 6.56
Get precalculated fractions of proteins
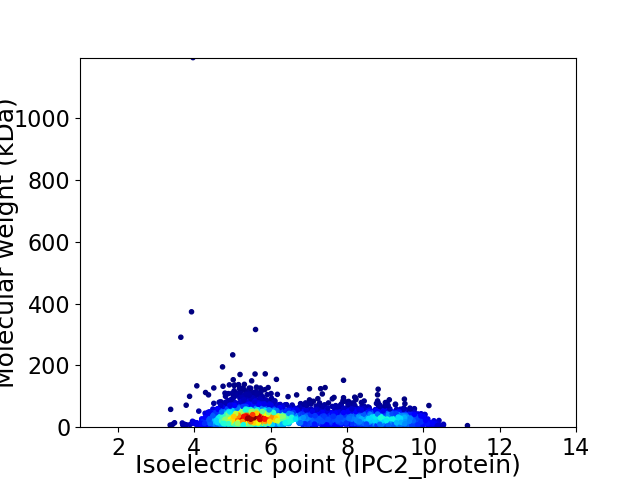
Virtual 2D-PAGE plot for 4486 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5B8B120|A0A5B8B120_9RHIZ Uncharacterized protein OS=Mesorhizobium sp. 8 OX=2584466 GN=FGU64_07890 PE=4 SV=1
MM1 pKa = 7.9RR2 pKa = 11.84DD3 pKa = 3.42DD4 pKa = 3.76YY5 pKa = 12.01LEE7 pKa = 4.51FKK9 pKa = 10.61ALQATSEE16 pKa = 4.05PTQADD21 pKa = 3.3DD22 pKa = 3.73HH23 pKa = 6.56AQLSFQSPYY32 pKa = 11.41DD33 pKa = 3.82FDD35 pKa = 6.59DD36 pKa = 5.0PDD38 pKa = 5.22PGIHH42 pKa = 5.79YY43 pKa = 10.33VPAGPTIEE51 pKa = 4.28TMTVPGGVAFTPMEE65 pKa = 4.33IPVVPAANPALYY77 pKa = 10.1VPSEE81 pKa = 4.34SYY83 pKa = 10.54PDD85 pKa = 4.48FISRR89 pKa = 11.84HH90 pKa = 4.09FTFAHH95 pKa = 5.88SSGSIQLQPPGSVATHH111 pKa = 5.16VVQINTLSDD120 pKa = 3.28NDD122 pKa = 3.88FVNVGAGNVTFHH134 pKa = 7.49AIGMPNLDD142 pKa = 4.92LDD144 pKa = 4.9ALYY147 pKa = 10.72QDD149 pKa = 4.93ALQFLPIPPASFSSTEE165 pKa = 4.27DD166 pKa = 2.44IGSFVNEE173 pKa = 3.8TAASLKK179 pKa = 10.78AFVAGVEE186 pKa = 4.22AAGQGTPDD194 pKa = 3.53GGAEE198 pKa = 3.94MSVLTDD204 pKa = 3.64DD205 pKa = 5.14GGVSVQTHH213 pKa = 5.14VSVFDD218 pKa = 4.01APIADD223 pKa = 4.94AIYY226 pKa = 10.16VGGRR230 pKa = 11.84IAAEE234 pKa = 3.95APKK237 pKa = 10.54LDD239 pKa = 4.3DD240 pKa = 4.03VLPAASPLVKK250 pKa = 10.6DD251 pKa = 4.07PVQAPSSADD260 pKa = 3.36PAGGNHH266 pKa = 7.47LIDD269 pKa = 4.53GSGSTATGVVIHH281 pKa = 6.03GTGNVAGEE289 pKa = 4.35TSVSLNTGSNALVNSATIVNDD310 pKa = 3.6ALEE313 pKa = 4.55GSVFAVAGNHH323 pKa = 5.24YY324 pKa = 10.01ALNAIVQVNAWSDD337 pKa = 3.23SDD339 pKa = 4.66SIGGSLSGWNAQQQPDD355 pKa = 3.64TTAFNIAQMLNIDD368 pKa = 3.66TGEE371 pKa = 4.07NAGSGNSGQPSFPKK385 pKa = 9.8AWAVTEE391 pKa = 4.05INGDD395 pKa = 4.31FISLNWTHH403 pKa = 5.89QVNFLLDD410 pKa = 3.28NDD412 pKa = 4.46TAVVSSTSGVTTTVGTGTNQQTNALSIADD441 pKa = 3.75LGKK444 pKa = 10.98YY445 pKa = 9.59YY446 pKa = 10.83DD447 pKa = 3.99LVLIGGSYY455 pKa = 10.67YY456 pKa = 10.57DD457 pKa = 4.44ANIITQTNVLLDD469 pKa = 3.73NDD471 pKa = 4.16VVGALANFQTQGQGSVSNGDD491 pKa = 3.25NLLWNNAKK499 pKa = 8.46ITSIGSSANAALPIGFAKK517 pKa = 10.22TLDD520 pKa = 4.0DD521 pKa = 4.22FAHH524 pKa = 6.61GNHH527 pKa = 6.3TLSSDD532 pKa = 3.83VLNDD536 pKa = 3.2EE537 pKa = 4.83AFQGLGGLKK546 pKa = 9.84VLYY549 pKa = 10.12ISGSVYY555 pKa = 10.35DD556 pKa = 4.36LQYY559 pKa = 10.92ISQTNVLGDD568 pKa = 3.74SDD570 pKa = 5.25DD571 pKa = 3.64VALALNAAHH580 pKa = 7.3ANTDD584 pKa = 3.98ANWSIVTGSNALINSAQIVDD604 pKa = 3.82VDD606 pKa = 4.32PAAKK610 pKa = 9.32TYY612 pKa = 11.08VGGDD616 pKa = 3.38HH617 pKa = 6.96YY618 pKa = 11.48SDD620 pKa = 3.96EE621 pKa = 5.16LLVQTDD627 pKa = 4.58IISTDD632 pKa = 3.44HH633 pKa = 7.32LLQTQDD639 pKa = 3.1ADD641 pKa = 3.79HH642 pKa = 6.69LVNEE646 pKa = 4.72AVAFLSDD653 pKa = 4.59DD654 pKa = 3.56MTGPAHH660 pKa = 6.76EE661 pKa = 5.06NPASQLQAPLDD672 pKa = 3.7VHH674 pKa = 6.59PAHH677 pKa = 6.88SDD679 pKa = 3.08VMQSVISS686 pKa = 3.74
MM1 pKa = 7.9RR2 pKa = 11.84DD3 pKa = 3.42DD4 pKa = 3.76YY5 pKa = 12.01LEE7 pKa = 4.51FKK9 pKa = 10.61ALQATSEE16 pKa = 4.05PTQADD21 pKa = 3.3DD22 pKa = 3.73HH23 pKa = 6.56AQLSFQSPYY32 pKa = 11.41DD33 pKa = 3.82FDD35 pKa = 6.59DD36 pKa = 5.0PDD38 pKa = 5.22PGIHH42 pKa = 5.79YY43 pKa = 10.33VPAGPTIEE51 pKa = 4.28TMTVPGGVAFTPMEE65 pKa = 4.33IPVVPAANPALYY77 pKa = 10.1VPSEE81 pKa = 4.34SYY83 pKa = 10.54PDD85 pKa = 4.48FISRR89 pKa = 11.84HH90 pKa = 4.09FTFAHH95 pKa = 5.88SSGSIQLQPPGSVATHH111 pKa = 5.16VVQINTLSDD120 pKa = 3.28NDD122 pKa = 3.88FVNVGAGNVTFHH134 pKa = 7.49AIGMPNLDD142 pKa = 4.92LDD144 pKa = 4.9ALYY147 pKa = 10.72QDD149 pKa = 4.93ALQFLPIPPASFSSTEE165 pKa = 4.27DD166 pKa = 2.44IGSFVNEE173 pKa = 3.8TAASLKK179 pKa = 10.78AFVAGVEE186 pKa = 4.22AAGQGTPDD194 pKa = 3.53GGAEE198 pKa = 3.94MSVLTDD204 pKa = 3.64DD205 pKa = 5.14GGVSVQTHH213 pKa = 5.14VSVFDD218 pKa = 4.01APIADD223 pKa = 4.94AIYY226 pKa = 10.16VGGRR230 pKa = 11.84IAAEE234 pKa = 3.95APKK237 pKa = 10.54LDD239 pKa = 4.3DD240 pKa = 4.03VLPAASPLVKK250 pKa = 10.6DD251 pKa = 4.07PVQAPSSADD260 pKa = 3.36PAGGNHH266 pKa = 7.47LIDD269 pKa = 4.53GSGSTATGVVIHH281 pKa = 6.03GTGNVAGEE289 pKa = 4.35TSVSLNTGSNALVNSATIVNDD310 pKa = 3.6ALEE313 pKa = 4.55GSVFAVAGNHH323 pKa = 5.24YY324 pKa = 10.01ALNAIVQVNAWSDD337 pKa = 3.23SDD339 pKa = 4.66SIGGSLSGWNAQQQPDD355 pKa = 3.64TTAFNIAQMLNIDD368 pKa = 3.66TGEE371 pKa = 4.07NAGSGNSGQPSFPKK385 pKa = 9.8AWAVTEE391 pKa = 4.05INGDD395 pKa = 4.31FISLNWTHH403 pKa = 5.89QVNFLLDD410 pKa = 3.28NDD412 pKa = 4.46TAVVSSTSGVTTTVGTGTNQQTNALSIADD441 pKa = 3.75LGKK444 pKa = 10.98YY445 pKa = 9.59YY446 pKa = 10.83DD447 pKa = 3.99LVLIGGSYY455 pKa = 10.67YY456 pKa = 10.57DD457 pKa = 4.44ANIITQTNVLLDD469 pKa = 3.73NDD471 pKa = 4.16VVGALANFQTQGQGSVSNGDD491 pKa = 3.25NLLWNNAKK499 pKa = 8.46ITSIGSSANAALPIGFAKK517 pKa = 10.22TLDD520 pKa = 4.0DD521 pKa = 4.22FAHH524 pKa = 6.61GNHH527 pKa = 6.3TLSSDD532 pKa = 3.83VLNDD536 pKa = 3.2EE537 pKa = 4.83AFQGLGGLKK546 pKa = 9.84VLYY549 pKa = 10.12ISGSVYY555 pKa = 10.35DD556 pKa = 4.36LQYY559 pKa = 10.92ISQTNVLGDD568 pKa = 3.74SDD570 pKa = 5.25DD571 pKa = 3.64VALALNAAHH580 pKa = 7.3ANTDD584 pKa = 3.98ANWSIVTGSNALINSAQIVDD604 pKa = 3.82VDD606 pKa = 4.32PAAKK610 pKa = 9.32TYY612 pKa = 11.08VGGDD616 pKa = 3.38HH617 pKa = 6.96YY618 pKa = 11.48SDD620 pKa = 3.96EE621 pKa = 5.16LLVQTDD627 pKa = 4.58IISTDD632 pKa = 3.44HH633 pKa = 7.32LLQTQDD639 pKa = 3.1ADD641 pKa = 3.79HH642 pKa = 6.69LVNEE646 pKa = 4.72AVAFLSDD653 pKa = 4.59DD654 pKa = 3.56MTGPAHH660 pKa = 6.76EE661 pKa = 5.06NPASQLQAPLDD672 pKa = 3.7VHH674 pKa = 6.59PAHH677 pKa = 6.88SDD679 pKa = 3.08VMQSVISS686 pKa = 3.74
Molecular weight: 71.22 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5B8B6G4|A0A5B8B6G4_9RHIZ ABC transporter ATP-binding protein OS=Mesorhizobium sp. 8 OX=2584466 GN=FGU64_16800 PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.25GGRR28 pKa = 11.84GVIAARR34 pKa = 11.84RR35 pKa = 11.84NRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.25GGRR28 pKa = 11.84GVIAARR34 pKa = 11.84RR35 pKa = 11.84NRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1410974 |
17 |
11922 |
314.5 |
34.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.929 ± 0.05 | 0.78 ± 0.012 |
5.752 ± 0.038 | 5.643 ± 0.041 |
3.852 ± 0.024 | 8.869 ± 0.06 |
1.961 ± 0.02 | 5.103 ± 0.029 |
3.607 ± 0.037 | 9.848 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.514 ± 0.026 | 2.565 ± 0.028 |
5.046 ± 0.036 | 2.877 ± 0.018 |
6.892 ± 0.053 | 5.261 ± 0.039 |
5.276 ± 0.053 | 7.661 ± 0.029 |
1.304 ± 0.016 | 2.259 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
