
Methanimicrococcus blatticola
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Methanomicrobia; Methanosarcinales; Methanosarcinaceae; Methanimicrococcus
Average proteome isoelectric point is 5.78
Get precalculated fractions of proteins
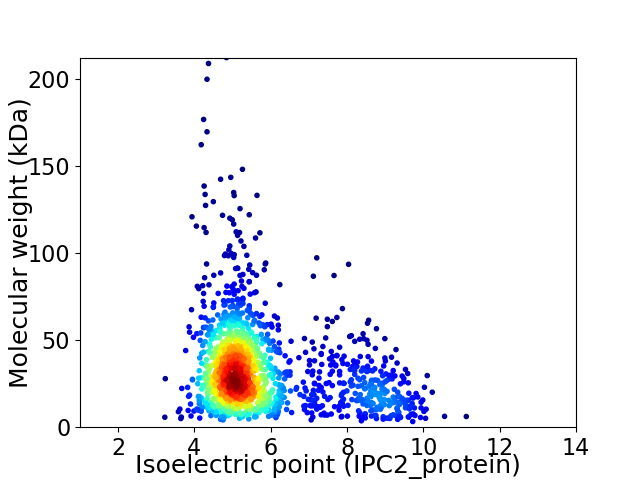
Virtual 2D-PAGE plot for 1537 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
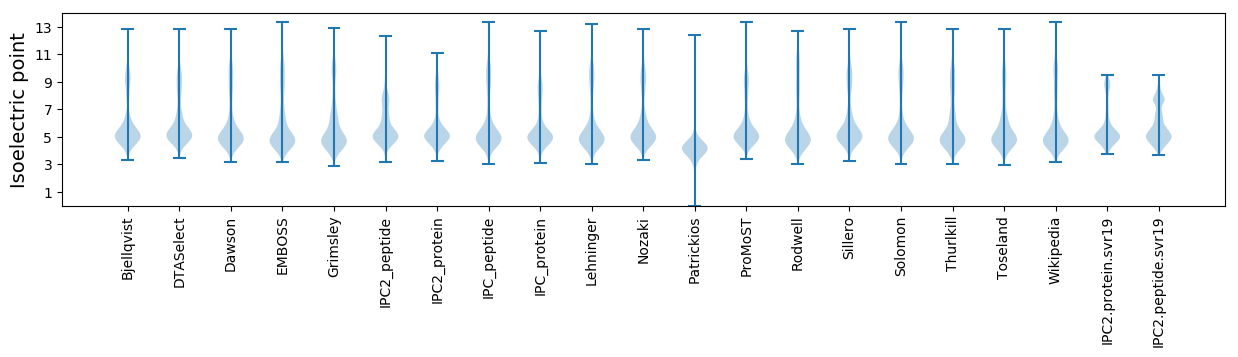
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A484F462|A0A484F462_9EURY Tritrans polycis-undecaprenyl-diphosphate synthase (geranylgeranyl-diphosphate specific) OS=Methanimicrococcus blatticola OX=91560 GN=uppS PE=3 SV=1
MM1 pKa = 7.32SASDD5 pKa = 3.64KK6 pKa = 11.09KK7 pKa = 11.29GGFIDD12 pKa = 4.21SKK14 pKa = 10.66YY15 pKa = 10.56LNIVTAVFVVFAVLLVCILMYY36 pKa = 10.7NPSLLGIQSASAHH49 pKa = 6.86PDD51 pKa = 3.36YY52 pKa = 10.81EE53 pKa = 4.56DD54 pKa = 4.92KK55 pKa = 11.01IFNQSMITSISIDD68 pKa = 3.26MSEE71 pKa = 4.47EE72 pKa = 3.11NWMALLEE79 pKa = 4.43NPLTEE84 pKa = 4.5EE85 pKa = 4.45FYY87 pKa = 10.4MANVTINGEE96 pKa = 4.08TFYY99 pKa = 11.42SVGIRR104 pKa = 11.84TKK106 pKa = 11.12GMTSLSQVASSDD118 pKa = 3.05SDD120 pKa = 3.44RR121 pKa = 11.84YY122 pKa = 10.49SFKK125 pKa = 11.04LKK127 pKa = 10.48ADD129 pKa = 3.74TYY131 pKa = 11.09IDD133 pKa = 3.6GQTFFGLEE141 pKa = 3.73EE142 pKa = 3.8FVINNMYY149 pKa = 10.45QDD151 pKa = 3.62PSYY154 pKa = 10.08MKK156 pKa = 10.21EE157 pKa = 3.73YY158 pKa = 10.27LAYY161 pKa = 10.94DD162 pKa = 3.4MMTYY166 pKa = 9.77MGVASPLFNFADD178 pKa = 2.96ITMNGEE184 pKa = 3.55PWGLYY189 pKa = 9.68LAIEE193 pKa = 4.25VMEE196 pKa = 4.8EE197 pKa = 3.74DD198 pKa = 3.28FAMRR202 pKa = 11.84VYY204 pKa = 10.98GSDD207 pKa = 3.18FGQLYY212 pKa = 10.09KK213 pKa = 10.68PEE215 pKa = 4.01TMGMGGGNVGGNRR228 pKa = 11.84PEE230 pKa = 4.21GGGNFGGGMGITRR243 pKa = 11.84PEE245 pKa = 4.57DD246 pKa = 3.37GDD248 pKa = 3.7FDD250 pKa = 5.42GDD252 pKa = 3.52FGEE255 pKa = 4.23ISNRR259 pKa = 11.84NGQMIPIQGNQAMPTASITLATSADD284 pKa = 3.61NQTITVNGTTFEE296 pKa = 4.21TQEE299 pKa = 4.53DD300 pKa = 4.8GTFTEE305 pKa = 4.52EE306 pKa = 4.15DD307 pKa = 3.27RR308 pKa = 11.84AAIAAAMAEE317 pKa = 4.24AGNTRR322 pKa = 11.84QNFGGMGTGGGADD335 pKa = 4.14LVYY338 pKa = 10.49TDD340 pKa = 5.81DD341 pKa = 5.01NISSYY346 pKa = 10.26SQIFEE351 pKa = 3.9NAVFKK356 pKa = 10.63NAKK359 pKa = 9.7NSDD362 pKa = 3.7FNRR365 pKa = 11.84VITALKK371 pKa = 9.12YY372 pKa = 10.43LNAGEE377 pKa = 4.2EE378 pKa = 4.4LEE380 pKa = 4.55TYY382 pKa = 10.95VDD384 pKa = 3.22VDD386 pKa = 3.05HH387 pKa = 6.89TLRR390 pKa = 11.84YY391 pKa = 8.76FAVNTVVVNLDD402 pKa = 3.87SYY404 pKa = 11.98VSGLKK409 pKa = 10.11HH410 pKa = 6.6NYY412 pKa = 9.25YY413 pKa = 10.5LYY415 pKa = 10.79EE416 pKa = 5.45KK417 pKa = 10.07DD418 pKa = 3.51GQITILPWDD427 pKa = 3.67YY428 pKa = 11.59NLAFGAFQSGTASSTVNFPIDD449 pKa = 3.7TPVSGVSMEE458 pKa = 4.13DD459 pKa = 3.05RR460 pKa = 11.84PLIGVLLAEE469 pKa = 4.77DD470 pKa = 4.75EE471 pKa = 4.49YY472 pKa = 11.2MDD474 pKa = 5.62LYY476 pKa = 11.06HH477 pKa = 7.59EE478 pKa = 4.27YY479 pKa = 10.28LQEE482 pKa = 4.48IVDD485 pKa = 4.11EE486 pKa = 4.6YY487 pKa = 11.3FNSGYY492 pKa = 10.21YY493 pKa = 10.1DD494 pKa = 3.54NKK496 pKa = 9.52ITEE499 pKa = 4.6LDD501 pKa = 3.76DD502 pKa = 5.78LIGEE506 pKa = 4.41HH507 pKa = 6.22VKK509 pKa = 10.77NDD511 pKa = 3.09QTAFFTYY518 pKa = 10.22DD519 pKa = 3.11EE520 pKa = 4.63YY521 pKa = 10.77VTAVEE526 pKa = 4.1EE527 pKa = 4.01MRR529 pKa = 11.84VFGEE533 pKa = 4.18LRR535 pKa = 11.84AEE537 pKa = 4.5SIQGQLDD544 pKa = 3.52GTVPSTSEE552 pKa = 4.0DD553 pKa = 3.18QTANPDD559 pKa = 3.65ALIDD563 pKa = 3.87ASALNMSALGGMGGGMGGGMPGGGGRR589 pKa = 11.84MQGGNWPTQVTTPQAADD606 pKa = 4.01DD607 pKa = 4.52LSVPPTGTGAAPAA620 pKa = 4.28
MM1 pKa = 7.32SASDD5 pKa = 3.64KK6 pKa = 11.09KK7 pKa = 11.29GGFIDD12 pKa = 4.21SKK14 pKa = 10.66YY15 pKa = 10.56LNIVTAVFVVFAVLLVCILMYY36 pKa = 10.7NPSLLGIQSASAHH49 pKa = 6.86PDD51 pKa = 3.36YY52 pKa = 10.81EE53 pKa = 4.56DD54 pKa = 4.92KK55 pKa = 11.01IFNQSMITSISIDD68 pKa = 3.26MSEE71 pKa = 4.47EE72 pKa = 3.11NWMALLEE79 pKa = 4.43NPLTEE84 pKa = 4.5EE85 pKa = 4.45FYY87 pKa = 10.4MANVTINGEE96 pKa = 4.08TFYY99 pKa = 11.42SVGIRR104 pKa = 11.84TKK106 pKa = 11.12GMTSLSQVASSDD118 pKa = 3.05SDD120 pKa = 3.44RR121 pKa = 11.84YY122 pKa = 10.49SFKK125 pKa = 11.04LKK127 pKa = 10.48ADD129 pKa = 3.74TYY131 pKa = 11.09IDD133 pKa = 3.6GQTFFGLEE141 pKa = 3.73EE142 pKa = 3.8FVINNMYY149 pKa = 10.45QDD151 pKa = 3.62PSYY154 pKa = 10.08MKK156 pKa = 10.21EE157 pKa = 3.73YY158 pKa = 10.27LAYY161 pKa = 10.94DD162 pKa = 3.4MMTYY166 pKa = 9.77MGVASPLFNFADD178 pKa = 2.96ITMNGEE184 pKa = 3.55PWGLYY189 pKa = 9.68LAIEE193 pKa = 4.25VMEE196 pKa = 4.8EE197 pKa = 3.74DD198 pKa = 3.28FAMRR202 pKa = 11.84VYY204 pKa = 10.98GSDD207 pKa = 3.18FGQLYY212 pKa = 10.09KK213 pKa = 10.68PEE215 pKa = 4.01TMGMGGGNVGGNRR228 pKa = 11.84PEE230 pKa = 4.21GGGNFGGGMGITRR243 pKa = 11.84PEE245 pKa = 4.57DD246 pKa = 3.37GDD248 pKa = 3.7FDD250 pKa = 5.42GDD252 pKa = 3.52FGEE255 pKa = 4.23ISNRR259 pKa = 11.84NGQMIPIQGNQAMPTASITLATSADD284 pKa = 3.61NQTITVNGTTFEE296 pKa = 4.21TQEE299 pKa = 4.53DD300 pKa = 4.8GTFTEE305 pKa = 4.52EE306 pKa = 4.15DD307 pKa = 3.27RR308 pKa = 11.84AAIAAAMAEE317 pKa = 4.24AGNTRR322 pKa = 11.84QNFGGMGTGGGADD335 pKa = 4.14LVYY338 pKa = 10.49TDD340 pKa = 5.81DD341 pKa = 5.01NISSYY346 pKa = 10.26SQIFEE351 pKa = 3.9NAVFKK356 pKa = 10.63NAKK359 pKa = 9.7NSDD362 pKa = 3.7FNRR365 pKa = 11.84VITALKK371 pKa = 9.12YY372 pKa = 10.43LNAGEE377 pKa = 4.2EE378 pKa = 4.4LEE380 pKa = 4.55TYY382 pKa = 10.95VDD384 pKa = 3.22VDD386 pKa = 3.05HH387 pKa = 6.89TLRR390 pKa = 11.84YY391 pKa = 8.76FAVNTVVVNLDD402 pKa = 3.87SYY404 pKa = 11.98VSGLKK409 pKa = 10.11HH410 pKa = 6.6NYY412 pKa = 9.25YY413 pKa = 10.5LYY415 pKa = 10.79EE416 pKa = 5.45KK417 pKa = 10.07DD418 pKa = 3.51GQITILPWDD427 pKa = 3.67YY428 pKa = 11.59NLAFGAFQSGTASSTVNFPIDD449 pKa = 3.7TPVSGVSMEE458 pKa = 4.13DD459 pKa = 3.05RR460 pKa = 11.84PLIGVLLAEE469 pKa = 4.77DD470 pKa = 4.75EE471 pKa = 4.49YY472 pKa = 11.2MDD474 pKa = 5.62LYY476 pKa = 11.06HH477 pKa = 7.59EE478 pKa = 4.27YY479 pKa = 10.28LQEE482 pKa = 4.48IVDD485 pKa = 4.11EE486 pKa = 4.6YY487 pKa = 11.3FNSGYY492 pKa = 10.21YY493 pKa = 10.1DD494 pKa = 3.54NKK496 pKa = 9.52ITEE499 pKa = 4.6LDD501 pKa = 3.76DD502 pKa = 5.78LIGEE506 pKa = 4.41HH507 pKa = 6.22VKK509 pKa = 10.77NDD511 pKa = 3.09QTAFFTYY518 pKa = 10.22DD519 pKa = 3.11EE520 pKa = 4.63YY521 pKa = 10.77VTAVEE526 pKa = 4.1EE527 pKa = 4.01MRR529 pKa = 11.84VFGEE533 pKa = 4.18LRR535 pKa = 11.84AEE537 pKa = 4.5SIQGQLDD544 pKa = 3.52GTVPSTSEE552 pKa = 4.0DD553 pKa = 3.18QTANPDD559 pKa = 3.65ALIDD563 pKa = 3.87ASALNMSALGGMGGGMGGGMPGGGGRR589 pKa = 11.84MQGGNWPTQVTTPQAADD606 pKa = 4.01DD607 pKa = 4.52LSVPPTGTGAAPAA620 pKa = 4.28
Molecular weight: 67.46 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A484F3W2|A0A484F3W2_9EURY UPF0219 protein C7391_1026 OS=Methanimicrococcus blatticola OX=91560 GN=C7391_1026 PE=3 SV=1
MM1 pKa = 7.51SKK3 pKa = 10.36NLKK6 pKa = 8.69GQKK9 pKa = 9.41KK10 pKa = 9.15RR11 pKa = 11.84LAKK14 pKa = 10.55AHH16 pKa = 5.14VQNARR21 pKa = 11.84VPVWVIIKK29 pKa = 7.41TKK31 pKa = 10.61RR32 pKa = 11.84NVTSHH37 pKa = 6.76PKK39 pKa = 8.23RR40 pKa = 11.84RR41 pKa = 11.84HH42 pKa = 3.63WRR44 pKa = 11.84RR45 pKa = 11.84SSMRR49 pKa = 11.84LKK51 pKa = 10.83
MM1 pKa = 7.51SKK3 pKa = 10.36NLKK6 pKa = 8.69GQKK9 pKa = 9.41KK10 pKa = 9.15RR11 pKa = 11.84LAKK14 pKa = 10.55AHH16 pKa = 5.14VQNARR21 pKa = 11.84VPVWVIIKK29 pKa = 7.41TKK31 pKa = 10.61RR32 pKa = 11.84NVTSHH37 pKa = 6.76PKK39 pKa = 8.23RR40 pKa = 11.84RR41 pKa = 11.84HH42 pKa = 3.63WRR44 pKa = 11.84RR45 pKa = 11.84SSMRR49 pKa = 11.84LKK51 pKa = 10.83
Molecular weight: 6.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
476405 |
25 |
1890 |
310.0 |
34.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.749 ± 0.07 | 1.186 ± 0.034 |
6.068 ± 0.053 | 7.481 ± 0.075 |
4.286 ± 0.05 | 7.217 ± 0.074 |
1.54 ± 0.025 | 7.896 ± 0.059 |
6.994 ± 0.057 | 8.412 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.742 ± 0.035 | 4.587 ± 0.064 |
3.879 ± 0.04 | 2.547 ± 0.031 |
3.767 ± 0.053 | 6.432 ± 0.065 |
5.83 ± 0.069 | 7.074 ± 0.06 |
0.789 ± 0.024 | 3.522 ± 0.042 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
