
Shinobi tetravirus
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified ssRNA viruses
Average proteome isoelectric point is 8.0
Get precalculated fractions of proteins
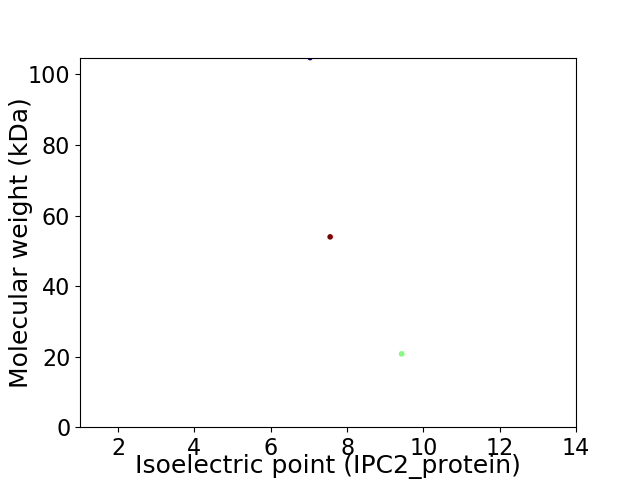
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
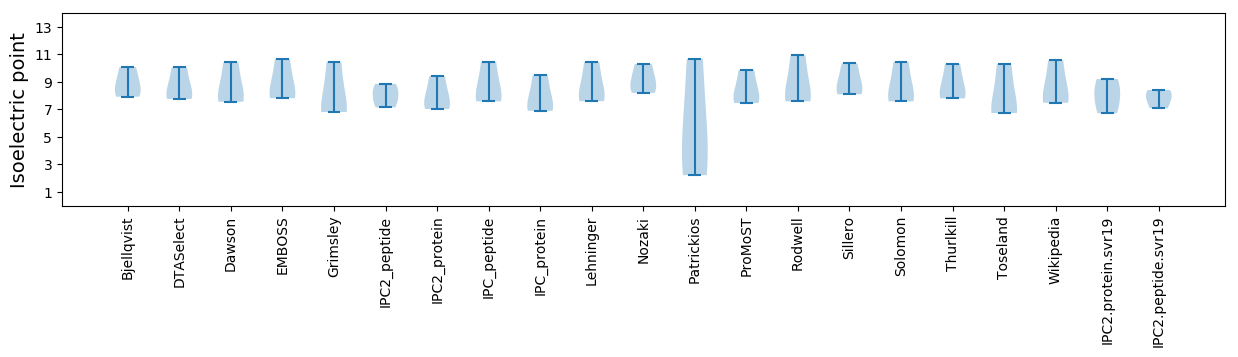
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A347ZKJ0|A0A347ZKJ0_9VIRU Hypothetical protein protein OS=Shinobi tetravirus OX=2006394 GN=hypothetical protein PE=4 SV=1
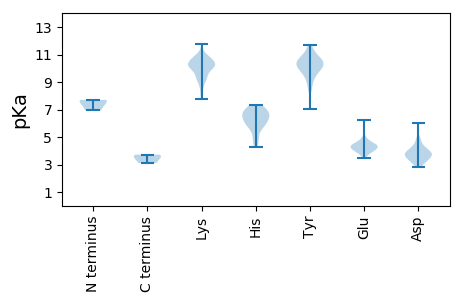
MM1 pKa = 6.94MNSVRR6 pKa = 11.84GFVTVNRR13 pKa = 11.84AFGNAAACDD22 pKa = 3.79LLLAEE27 pKa = 4.99GALTAFSHH35 pKa = 5.84EE36 pKa = 4.83EE37 pKa = 3.5IMSARR42 pKa = 11.84KK43 pKa = 9.63KK44 pKa = 10.21RR45 pKa = 11.84VYY47 pKa = 10.42CAGTHH52 pKa = 4.24KK53 pKa = 9.87TFVEE57 pKa = 4.38RR58 pKa = 11.84TKK60 pKa = 11.15LLLGRR65 pKa = 11.84KK66 pKa = 8.89CKK68 pKa = 10.49SPYY71 pKa = 9.91KK72 pKa = 8.93YY73 pKa = 9.1TVVAQKK79 pKa = 10.01LQKK82 pKa = 10.18FMPVGVLPDD91 pKa = 3.35WTDD94 pKa = 2.8VDD96 pKa = 4.2SLLEE100 pKa = 3.98RR101 pKa = 11.84VKK103 pKa = 9.79VTKK106 pKa = 10.41AAGAGAPYY114 pKa = 10.06WKK116 pKa = 10.33DD117 pKa = 2.88KK118 pKa = 11.2GDD120 pKa = 4.14AFDD123 pKa = 4.46EE124 pKa = 4.42CMQEE128 pKa = 4.38ILPTLIKK135 pKa = 10.44AINDD139 pKa = 3.57GTVEE143 pKa = 3.77QLARR147 pKa = 11.84GRR149 pKa = 11.84PEE151 pKa = 3.5WFLVEE156 pKa = 4.68LKK158 pKa = 11.04NKK160 pKa = 8.97IDD162 pKa = 3.81RR163 pKa = 11.84YY164 pKa = 9.58EE165 pKa = 4.02LPISKK170 pKa = 8.21TRR172 pKa = 11.84PYY174 pKa = 11.24GCFPFHH180 pKa = 5.94WTLLFSCLIQPFCEE194 pKa = 4.36ALSVWTDD201 pKa = 2.89SKK203 pKa = 10.2TCNAYY208 pKa = 10.16GMSFCSGNLKK218 pKa = 9.98KK219 pKa = 9.96WFSRR223 pKa = 11.84VEE225 pKa = 4.01SLIATAKK232 pKa = 10.34KK233 pKa = 10.04KK234 pKa = 10.72NKK236 pKa = 9.42DD237 pKa = 3.26MYY239 pKa = 10.87MMGVYY244 pKa = 10.57GDD246 pKa = 4.18DD247 pKa = 3.98CRR249 pKa = 11.84LIQAKK254 pKa = 10.2ASGGAVVEE262 pKa = 4.48DD263 pKa = 4.73PDD265 pKa = 3.95MKK267 pKa = 10.93QHH269 pKa = 7.16DD270 pKa = 4.25GAIDD274 pKa = 3.54TEE276 pKa = 4.85TISGTVKK283 pKa = 10.33WIYY286 pKa = 11.43DD287 pKa = 3.71MYY289 pKa = 11.13AKK291 pKa = 10.06QHH293 pKa = 6.2GDD295 pKa = 3.17SKK297 pKa = 10.7FWEE300 pKa = 4.75FVLQQLAAFACDD312 pKa = 3.51PNMVIDD318 pKa = 4.92GPTVYY323 pKa = 9.56TKK325 pKa = 10.39KK326 pKa = 10.79VPGGLISGVPGTTLFGCAKK345 pKa = 10.21SVLAYY350 pKa = 10.35AGLVDD355 pKa = 3.89EE356 pKa = 5.6CMFDD360 pKa = 2.94KK361 pKa = 10.95TLFLNEE367 pKa = 4.43AKK369 pKa = 10.6VKK371 pKa = 10.19AYY373 pKa = 10.84LMEE376 pKa = 4.09KK377 pKa = 9.82HH378 pKa = 6.11GCALKK383 pKa = 10.9DD384 pKa = 3.7GTWRR388 pKa = 11.84PSPMNLSQADD398 pKa = 3.72GTYY401 pKa = 10.11LCEE404 pKa = 4.45SKK406 pKa = 10.8FLGMRR411 pKa = 11.84LMWRR415 pKa = 11.84TSEE418 pKa = 4.0QFPEE422 pKa = 4.39PFLVPTLHH430 pKa = 7.03DD431 pKa = 4.41GEE433 pKa = 4.22WLEE436 pKa = 6.24LIMCPRR442 pKa = 11.84TSGDD446 pKa = 2.98KK447 pKa = 10.59KK448 pKa = 10.72GSHH451 pKa = 6.63LSEE454 pKa = 3.73QRR456 pKa = 11.84TSFDD460 pKa = 3.92RR461 pKa = 11.84IRR463 pKa = 11.84GLLLCGAVYY472 pKa = 10.03SEE474 pKa = 4.46RR475 pKa = 11.84ARR477 pKa = 11.84AALLAAVDD485 pKa = 4.64LVPATAVLLSTQLDD499 pKa = 3.74GRR501 pKa = 11.84KK502 pKa = 9.85GLIEE506 pKa = 4.42SPNDD510 pKa = 3.44LVVSEE515 pKa = 4.63DD516 pKa = 4.13FIYY519 pKa = 10.25PSTAGIPGLEE529 pKa = 4.3FCDD532 pKa = 3.58ALYY535 pKa = 11.01NLSAEE540 pKa = 4.09QRR542 pKa = 11.84EE543 pKa = 4.7GCKK546 pKa = 10.2FEE548 pKa = 4.87SVYY551 pKa = 9.29PTLDD555 pKa = 3.29DD556 pKa = 3.93FVNSRR561 pKa = 11.84KK562 pKa = 9.14EE563 pKa = 3.66LHH565 pKa = 6.53RR566 pKa = 11.84SRR568 pKa = 11.84FRR570 pKa = 11.84YY571 pKa = 9.55IIHH574 pKa = 6.33QSLPAKK580 pKa = 8.43PTAKK584 pKa = 10.1PIPAPRR590 pKa = 11.84SYY592 pKa = 8.43THH594 pKa = 6.68KK595 pKa = 10.0HH596 pKa = 4.92TGQQVFLPKK605 pKa = 9.85PAPRR609 pKa = 11.84LQTTSEE615 pKa = 4.0QPAMDD620 pKa = 4.51KK621 pKa = 10.41EE622 pKa = 4.53VQEE625 pKa = 4.14QEE627 pKa = 4.36PPLFTANEE635 pKa = 4.35PIVAVEE641 pKa = 4.3EE642 pKa = 4.44EE643 pKa = 4.52FTSEE647 pKa = 4.09DD648 pKa = 3.67LEE650 pKa = 4.89KK651 pKa = 10.6IPLPPIPEE659 pKa = 4.37PQHH662 pKa = 5.65GFARR666 pKa = 11.84GGRR669 pKa = 11.84EE670 pKa = 3.66VRR672 pKa = 11.84AGSLHH677 pKa = 6.52SKK679 pKa = 10.0DD680 pKa = 3.93GKK682 pKa = 9.37TIAPRR687 pKa = 11.84HH688 pKa = 5.13KK689 pKa = 9.87LQRR692 pKa = 11.84LEE694 pKa = 4.22TYY696 pKa = 10.35LGEE699 pKa = 4.31GLKK702 pKa = 10.69KK703 pKa = 9.85NAKK706 pKa = 9.32ALVMSNPVLPEE717 pKa = 3.75GTVRR721 pKa = 11.84NYY723 pKa = 10.28QCLFSAVLDD732 pKa = 4.41RR733 pKa = 11.84GFKK736 pKa = 9.31WPHH739 pKa = 6.53DD740 pKa = 4.25LPEE743 pKa = 4.53GTHH746 pKa = 7.1DD747 pKa = 3.75NALVGQFAAINGLKK761 pKa = 10.2SSPEE765 pKa = 3.78TSVIDD770 pKa = 3.78PAQDD774 pKa = 3.29MKK776 pKa = 10.76GVWMRR781 pKa = 11.84LVPKK785 pKa = 10.28DD786 pKa = 3.74GLGSSIDD793 pKa = 3.9YY794 pKa = 10.54AQLTATGCTLKK805 pKa = 10.73RR806 pKa = 11.84MQAAINQQFAAWLRR820 pKa = 11.84EE821 pKa = 3.92FKK823 pKa = 10.44EE824 pKa = 4.59SFMEE828 pKa = 4.27PPKK831 pKa = 10.68KK832 pKa = 10.35VIKK835 pKa = 9.42PPPGAPGFTSSNWAAEE851 pKa = 4.18VEE853 pKa = 4.21SDD855 pKa = 3.64DD856 pKa = 4.17LSRR859 pKa = 11.84MGSLSEE865 pKa = 4.28ASLKK869 pKa = 9.97EE870 pKa = 3.91VQRR873 pKa = 11.84VVRR876 pKa = 11.84DD877 pKa = 3.34LLRR880 pKa = 11.84TEE882 pKa = 4.52FAYY885 pKa = 10.79LRR887 pKa = 11.84TLDD890 pKa = 3.87RR891 pKa = 11.84EE892 pKa = 4.25HH893 pKa = 6.88TSRR896 pKa = 11.84HH897 pKa = 4.47GTKK900 pKa = 9.32QQHH903 pKa = 5.91KK904 pKa = 8.75VSKK907 pKa = 10.65SPASPTNAEE916 pKa = 3.85EE917 pKa = 4.2EE918 pKa = 4.53GRR920 pKa = 11.84SEE922 pKa = 4.28GRR924 pKa = 11.84DD925 pKa = 3.3ISPVEE930 pKa = 3.97ASPSRR935 pKa = 11.84TVPSAGDD942 pKa = 3.26NN943 pKa = 3.53
MM1 pKa = 6.94MNSVRR6 pKa = 11.84GFVTVNRR13 pKa = 11.84AFGNAAACDD22 pKa = 3.79LLLAEE27 pKa = 4.99GALTAFSHH35 pKa = 5.84EE36 pKa = 4.83EE37 pKa = 3.5IMSARR42 pKa = 11.84KK43 pKa = 9.63KK44 pKa = 10.21RR45 pKa = 11.84VYY47 pKa = 10.42CAGTHH52 pKa = 4.24KK53 pKa = 9.87TFVEE57 pKa = 4.38RR58 pKa = 11.84TKK60 pKa = 11.15LLLGRR65 pKa = 11.84KK66 pKa = 8.89CKK68 pKa = 10.49SPYY71 pKa = 9.91KK72 pKa = 8.93YY73 pKa = 9.1TVVAQKK79 pKa = 10.01LQKK82 pKa = 10.18FMPVGVLPDD91 pKa = 3.35WTDD94 pKa = 2.8VDD96 pKa = 4.2SLLEE100 pKa = 3.98RR101 pKa = 11.84VKK103 pKa = 9.79VTKK106 pKa = 10.41AAGAGAPYY114 pKa = 10.06WKK116 pKa = 10.33DD117 pKa = 2.88KK118 pKa = 11.2GDD120 pKa = 4.14AFDD123 pKa = 4.46EE124 pKa = 4.42CMQEE128 pKa = 4.38ILPTLIKK135 pKa = 10.44AINDD139 pKa = 3.57GTVEE143 pKa = 3.77QLARR147 pKa = 11.84GRR149 pKa = 11.84PEE151 pKa = 3.5WFLVEE156 pKa = 4.68LKK158 pKa = 11.04NKK160 pKa = 8.97IDD162 pKa = 3.81RR163 pKa = 11.84YY164 pKa = 9.58EE165 pKa = 4.02LPISKK170 pKa = 8.21TRR172 pKa = 11.84PYY174 pKa = 11.24GCFPFHH180 pKa = 5.94WTLLFSCLIQPFCEE194 pKa = 4.36ALSVWTDD201 pKa = 2.89SKK203 pKa = 10.2TCNAYY208 pKa = 10.16GMSFCSGNLKK218 pKa = 9.98KK219 pKa = 9.96WFSRR223 pKa = 11.84VEE225 pKa = 4.01SLIATAKK232 pKa = 10.34KK233 pKa = 10.04KK234 pKa = 10.72NKK236 pKa = 9.42DD237 pKa = 3.26MYY239 pKa = 10.87MMGVYY244 pKa = 10.57GDD246 pKa = 4.18DD247 pKa = 3.98CRR249 pKa = 11.84LIQAKK254 pKa = 10.2ASGGAVVEE262 pKa = 4.48DD263 pKa = 4.73PDD265 pKa = 3.95MKK267 pKa = 10.93QHH269 pKa = 7.16DD270 pKa = 4.25GAIDD274 pKa = 3.54TEE276 pKa = 4.85TISGTVKK283 pKa = 10.33WIYY286 pKa = 11.43DD287 pKa = 3.71MYY289 pKa = 11.13AKK291 pKa = 10.06QHH293 pKa = 6.2GDD295 pKa = 3.17SKK297 pKa = 10.7FWEE300 pKa = 4.75FVLQQLAAFACDD312 pKa = 3.51PNMVIDD318 pKa = 4.92GPTVYY323 pKa = 9.56TKK325 pKa = 10.39KK326 pKa = 10.79VPGGLISGVPGTTLFGCAKK345 pKa = 10.21SVLAYY350 pKa = 10.35AGLVDD355 pKa = 3.89EE356 pKa = 5.6CMFDD360 pKa = 2.94KK361 pKa = 10.95TLFLNEE367 pKa = 4.43AKK369 pKa = 10.6VKK371 pKa = 10.19AYY373 pKa = 10.84LMEE376 pKa = 4.09KK377 pKa = 9.82HH378 pKa = 6.11GCALKK383 pKa = 10.9DD384 pKa = 3.7GTWRR388 pKa = 11.84PSPMNLSQADD398 pKa = 3.72GTYY401 pKa = 10.11LCEE404 pKa = 4.45SKK406 pKa = 10.8FLGMRR411 pKa = 11.84LMWRR415 pKa = 11.84TSEE418 pKa = 4.0QFPEE422 pKa = 4.39PFLVPTLHH430 pKa = 7.03DD431 pKa = 4.41GEE433 pKa = 4.22WLEE436 pKa = 6.24LIMCPRR442 pKa = 11.84TSGDD446 pKa = 2.98KK447 pKa = 10.59KK448 pKa = 10.72GSHH451 pKa = 6.63LSEE454 pKa = 3.73QRR456 pKa = 11.84TSFDD460 pKa = 3.92RR461 pKa = 11.84IRR463 pKa = 11.84GLLLCGAVYY472 pKa = 10.03SEE474 pKa = 4.46RR475 pKa = 11.84ARR477 pKa = 11.84AALLAAVDD485 pKa = 4.64LVPATAVLLSTQLDD499 pKa = 3.74GRR501 pKa = 11.84KK502 pKa = 9.85GLIEE506 pKa = 4.42SPNDD510 pKa = 3.44LVVSEE515 pKa = 4.63DD516 pKa = 4.13FIYY519 pKa = 10.25PSTAGIPGLEE529 pKa = 4.3FCDD532 pKa = 3.58ALYY535 pKa = 11.01NLSAEE540 pKa = 4.09QRR542 pKa = 11.84EE543 pKa = 4.7GCKK546 pKa = 10.2FEE548 pKa = 4.87SVYY551 pKa = 9.29PTLDD555 pKa = 3.29DD556 pKa = 3.93FVNSRR561 pKa = 11.84KK562 pKa = 9.14EE563 pKa = 3.66LHH565 pKa = 6.53RR566 pKa = 11.84SRR568 pKa = 11.84FRR570 pKa = 11.84YY571 pKa = 9.55IIHH574 pKa = 6.33QSLPAKK580 pKa = 8.43PTAKK584 pKa = 10.1PIPAPRR590 pKa = 11.84SYY592 pKa = 8.43THH594 pKa = 6.68KK595 pKa = 10.0HH596 pKa = 4.92TGQQVFLPKK605 pKa = 9.85PAPRR609 pKa = 11.84LQTTSEE615 pKa = 4.0QPAMDD620 pKa = 4.51KK621 pKa = 10.41EE622 pKa = 4.53VQEE625 pKa = 4.14QEE627 pKa = 4.36PPLFTANEE635 pKa = 4.35PIVAVEE641 pKa = 4.3EE642 pKa = 4.44EE643 pKa = 4.52FTSEE647 pKa = 4.09DD648 pKa = 3.67LEE650 pKa = 4.89KK651 pKa = 10.6IPLPPIPEE659 pKa = 4.37PQHH662 pKa = 5.65GFARR666 pKa = 11.84GGRR669 pKa = 11.84EE670 pKa = 3.66VRR672 pKa = 11.84AGSLHH677 pKa = 6.52SKK679 pKa = 10.0DD680 pKa = 3.93GKK682 pKa = 9.37TIAPRR687 pKa = 11.84HH688 pKa = 5.13KK689 pKa = 9.87LQRR692 pKa = 11.84LEE694 pKa = 4.22TYY696 pKa = 10.35LGEE699 pKa = 4.31GLKK702 pKa = 10.69KK703 pKa = 9.85NAKK706 pKa = 9.32ALVMSNPVLPEE717 pKa = 3.75GTVRR721 pKa = 11.84NYY723 pKa = 10.28QCLFSAVLDD732 pKa = 4.41RR733 pKa = 11.84GFKK736 pKa = 9.31WPHH739 pKa = 6.53DD740 pKa = 4.25LPEE743 pKa = 4.53GTHH746 pKa = 7.1DD747 pKa = 3.75NALVGQFAAINGLKK761 pKa = 10.2SSPEE765 pKa = 3.78TSVIDD770 pKa = 3.78PAQDD774 pKa = 3.29MKK776 pKa = 10.76GVWMRR781 pKa = 11.84LVPKK785 pKa = 10.28DD786 pKa = 3.74GLGSSIDD793 pKa = 3.9YY794 pKa = 10.54AQLTATGCTLKK805 pKa = 10.73RR806 pKa = 11.84MQAAINQQFAAWLRR820 pKa = 11.84EE821 pKa = 3.92FKK823 pKa = 10.44EE824 pKa = 4.59SFMEE828 pKa = 4.27PPKK831 pKa = 10.68KK832 pKa = 10.35VIKK835 pKa = 9.42PPPGAPGFTSSNWAAEE851 pKa = 4.18VEE853 pKa = 4.21SDD855 pKa = 3.64DD856 pKa = 4.17LSRR859 pKa = 11.84MGSLSEE865 pKa = 4.28ASLKK869 pKa = 9.97EE870 pKa = 3.91VQRR873 pKa = 11.84VVRR876 pKa = 11.84DD877 pKa = 3.34LLRR880 pKa = 11.84TEE882 pKa = 4.52FAYY885 pKa = 10.79LRR887 pKa = 11.84TLDD890 pKa = 3.87RR891 pKa = 11.84EE892 pKa = 4.25HH893 pKa = 6.88TSRR896 pKa = 11.84HH897 pKa = 4.47GTKK900 pKa = 9.32QQHH903 pKa = 5.91KK904 pKa = 8.75VSKK907 pKa = 10.65SPASPTNAEE916 pKa = 3.85EE917 pKa = 4.2EE918 pKa = 4.53GRR920 pKa = 11.84SEE922 pKa = 4.28GRR924 pKa = 11.84DD925 pKa = 3.3ISPVEE930 pKa = 3.97ASPSRR935 pKa = 11.84TVPSAGDD942 pKa = 3.26NN943 pKa = 3.53
Molecular weight: 104.78 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A347ZKJ1|A0A347ZKJ1_9VIRU Putative capsid protein protein OS=Shinobi tetravirus OX=2006394 GN=putative capsid protein PE=4 SV=1
MM1 pKa = 7.59ARR3 pKa = 11.84NNNIKK8 pKa = 10.3SASRR12 pKa = 11.84PQAQPTRR19 pKa = 11.84RR20 pKa = 11.84KK21 pKa = 9.61RR22 pKa = 11.84GGVKK26 pKa = 10.27DD27 pKa = 3.49VTSRR31 pKa = 11.84QWKK34 pKa = 8.5QVQAGLSRR42 pKa = 11.84ALEE45 pKa = 4.38IIRR48 pKa = 11.84SPLCFVPLILSIFLFYY64 pKa = 10.59DD65 pKa = 3.95DD66 pKa = 6.01KK67 pKa = 11.76LLTQLANTFSANAAFKK83 pKa = 10.27WLGTFISEE91 pKa = 4.55HH92 pKa = 6.21KK93 pKa = 10.22KK94 pKa = 10.58QIAGLIFLVPATFLSCDD111 pKa = 4.03PKK113 pKa = 10.3WATLVSAVVSYY124 pKa = 10.44LVSEE128 pKa = 4.33VFPAPVKK135 pKa = 9.14YY136 pKa = 10.28FEE138 pKa = 4.09YY139 pKa = 10.71GFLGGSLILLCTARR153 pKa = 11.84KK154 pKa = 8.98PIQFVVATVVALLTIGLGLWGSEE177 pKa = 4.11VFASLPSTSRR187 pKa = 11.84QGG189 pKa = 3.11
MM1 pKa = 7.59ARR3 pKa = 11.84NNNIKK8 pKa = 10.3SASRR12 pKa = 11.84PQAQPTRR19 pKa = 11.84RR20 pKa = 11.84KK21 pKa = 9.61RR22 pKa = 11.84GGVKK26 pKa = 10.27DD27 pKa = 3.49VTSRR31 pKa = 11.84QWKK34 pKa = 8.5QVQAGLSRR42 pKa = 11.84ALEE45 pKa = 4.38IIRR48 pKa = 11.84SPLCFVPLILSIFLFYY64 pKa = 10.59DD65 pKa = 3.95DD66 pKa = 6.01KK67 pKa = 11.76LLTQLANTFSANAAFKK83 pKa = 10.27WLGTFISEE91 pKa = 4.55HH92 pKa = 6.21KK93 pKa = 10.22KK94 pKa = 10.58QIAGLIFLVPATFLSCDD111 pKa = 4.03PKK113 pKa = 10.3WATLVSAVVSYY124 pKa = 10.44LVSEE128 pKa = 4.33VFPAPVKK135 pKa = 9.14YY136 pKa = 10.28FEE138 pKa = 4.09YY139 pKa = 10.71GFLGGSLILLCTARR153 pKa = 11.84KK154 pKa = 8.98PIQFVVATVVALLTIGLGLWGSEE177 pKa = 4.11VFASLPSTSRR187 pKa = 11.84QGG189 pKa = 3.11
Molecular weight: 20.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1627 |
189 |
943 |
542.3 |
59.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.498 ± 0.991 | 3.012 ± 0.838 |
4.364 ± 0.87 | 5.224 ± 1.296 |
4.364 ± 0.448 | 7.314 ± 0.388 |
2.274 ± 0.401 | 3.442 ± 0.436 |
5.778 ± 1.281 | 9.65 ± 0.537 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.028 ± 0.522 | 2.52 ± 0.038 |
7.376 ± 0.935 | 3.38 ± 0.384 |
5.286 ± 0.015 | 7.314 ± 0.393 |
7.253 ± 1.389 | 7.314 ± 0.978 |
1.782 ± 0.159 | 2.827 ± 0.242 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
