
Arcticibacterium luteifluviistationis
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Cytophagaceae; Arcticibacterium
Average proteome isoelectric point is 6.26
Get precalculated fractions of proteins
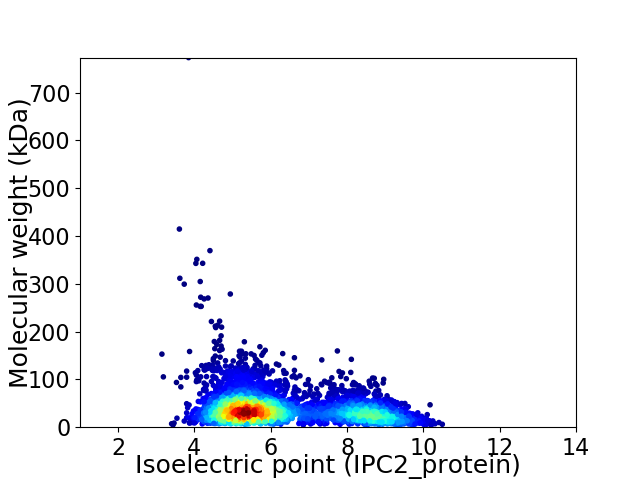
Virtual 2D-PAGE plot for 4329 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z4GB05|A0A2Z4GB05_9BACT Dehydrogenase OS=Arcticibacterium luteifluviistationis OX=1784714 GN=DJ013_09470 PE=4 SV=1
MM1 pKa = 7.12STLKK5 pKa = 10.19EE6 pKa = 4.1KK7 pKa = 11.06NMSNTLNKK15 pKa = 10.29LLLLAFTLFAFIHH28 pKa = 6.12EE29 pKa = 5.3GYY31 pKa = 9.25SQNNADD37 pKa = 4.12PGIGILMSPSIVIQGSTGILSATVGNYY64 pKa = 9.93GNEE67 pKa = 4.21TIVEE71 pKa = 3.92NSLRR75 pKa = 11.84VTITVGSNAEE85 pKa = 4.23IIGIAPGSDD94 pKa = 3.51SRR96 pKa = 11.84WSQLSLTTGSANSIKK111 pKa = 9.91LTNTVGGFNSFDD123 pKa = 3.37VGDD126 pKa = 3.6ILLTVRR132 pKa = 11.84GNLVSAPEE140 pKa = 4.73LILGNIVYY148 pKa = 8.49ITAEE152 pKa = 4.15NPLLCDD158 pKa = 4.19GCPSPPYY165 pKa = 10.26NVSQGNASSLNDD177 pKa = 3.37NSQTSLATSAPVIDD191 pKa = 4.29AVVDD195 pKa = 3.65ATAAVSGLTGGITSALTSNDD215 pKa = 3.39SLNGIVVVIGIDD227 pKa = 3.42SGNVTLTGLNVPAGLTLNADD247 pKa = 3.5GTVTVDD253 pKa = 3.18SNTPAGNYY261 pKa = 7.98NVEE264 pKa = 4.13YY265 pKa = 10.38KK266 pKa = 10.11ICEE269 pKa = 4.15VNNPTNCDD277 pKa = 3.32SVISMIVVSAPIIAAVVDD295 pKa = 3.84TTAAVSGLTGGITSALTSNDD315 pKa = 3.39SLNGIVVVIGIDD327 pKa = 3.24SGNVMLTGLTVPAGLTLNADD347 pKa = 3.5GTVTVDD353 pKa = 3.18SNTPAGNYY361 pKa = 7.98NVEE364 pKa = 4.13YY365 pKa = 10.38KK366 pKa = 10.11ICEE369 pKa = 4.15VNNPTNCDD377 pKa = 3.32SVISMIVVSAPIIAAVVDD395 pKa = 3.84TTAAVSGLTGGITSALTSNDD415 pKa = 3.39SLNGIVVVIGIDD427 pKa = 3.42SGNVTLTGLNVPAGLTLNADD447 pKa = 3.5GTVTVDD453 pKa = 3.18SNTPAGNYY461 pKa = 7.98NVEE464 pKa = 4.13YY465 pKa = 10.38KK466 pKa = 10.11ICEE469 pKa = 4.15VNNPTNCDD477 pKa = 3.32SVISMIVVSAPIIAAVVDD495 pKa = 3.84TTAAVSGLTGGITSALTSNDD515 pKa = 3.39SLNGIVVVIGIDD527 pKa = 3.24SGNVMLTGLTVPAGLTLNADD547 pKa = 3.5GTVTVDD553 pKa = 3.18SNTPAGNYY561 pKa = 7.98NVEE564 pKa = 4.13YY565 pKa = 10.38KK566 pKa = 10.11ICEE569 pKa = 4.15VNNPTNCDD577 pKa = 3.32SVISMIVVSAPIIAAVVDD595 pKa = 3.84TTAAVSGLTGGITSALTSNDD615 pKa = 3.39SLNGIVVVIGIDD627 pKa = 3.42SGNVTLTGLNVPAGLTLNADD647 pKa = 3.5GTVTVDD653 pKa = 3.18SNTPAGNYY661 pKa = 7.98NVEE664 pKa = 4.13YY665 pKa = 10.38KK666 pKa = 10.11ICEE669 pKa = 4.15VNNPTNCDD677 pKa = 3.3SVISMIEE684 pKa = 4.01VYY686 pKa = 10.36RR687 pKa = 11.84EE688 pKa = 4.03LPDD691 pKa = 3.88LSPTIDD697 pKa = 2.97IDD699 pKa = 4.03ALVFPIAGSAKK710 pKa = 10.29DD711 pKa = 3.52FVVNIGDD718 pKa = 3.8VKK720 pKa = 10.9GVEE723 pKa = 4.04SDD725 pKa = 3.52GQVVVKK731 pKa = 10.09VSKK734 pKa = 10.98GNAFFISYY742 pKa = 10.84ADD744 pKa = 3.45STSNSNANGGSVPVNNSAWIITEE767 pKa = 3.72NASFITITLKK777 pKa = 10.79PGVQISANTFSAIGFTITRR796 pKa = 11.84KK797 pKa = 10.31AGVPTQTSQPITVTIVNGTGLDD819 pKa = 3.75SQNSNNAYY827 pKa = 8.58STIVTAQQ834 pKa = 2.84
MM1 pKa = 7.12STLKK5 pKa = 10.19EE6 pKa = 4.1KK7 pKa = 11.06NMSNTLNKK15 pKa = 10.29LLLLAFTLFAFIHH28 pKa = 6.12EE29 pKa = 5.3GYY31 pKa = 9.25SQNNADD37 pKa = 4.12PGIGILMSPSIVIQGSTGILSATVGNYY64 pKa = 9.93GNEE67 pKa = 4.21TIVEE71 pKa = 3.92NSLRR75 pKa = 11.84VTITVGSNAEE85 pKa = 4.23IIGIAPGSDD94 pKa = 3.51SRR96 pKa = 11.84WSQLSLTTGSANSIKK111 pKa = 9.91LTNTVGGFNSFDD123 pKa = 3.37VGDD126 pKa = 3.6ILLTVRR132 pKa = 11.84GNLVSAPEE140 pKa = 4.73LILGNIVYY148 pKa = 8.49ITAEE152 pKa = 4.15NPLLCDD158 pKa = 4.19GCPSPPYY165 pKa = 10.26NVSQGNASSLNDD177 pKa = 3.37NSQTSLATSAPVIDD191 pKa = 4.29AVVDD195 pKa = 3.65ATAAVSGLTGGITSALTSNDD215 pKa = 3.39SLNGIVVVIGIDD227 pKa = 3.42SGNVTLTGLNVPAGLTLNADD247 pKa = 3.5GTVTVDD253 pKa = 3.18SNTPAGNYY261 pKa = 7.98NVEE264 pKa = 4.13YY265 pKa = 10.38KK266 pKa = 10.11ICEE269 pKa = 4.15VNNPTNCDD277 pKa = 3.32SVISMIVVSAPIIAAVVDD295 pKa = 3.84TTAAVSGLTGGITSALTSNDD315 pKa = 3.39SLNGIVVVIGIDD327 pKa = 3.24SGNVMLTGLTVPAGLTLNADD347 pKa = 3.5GTVTVDD353 pKa = 3.18SNTPAGNYY361 pKa = 7.98NVEE364 pKa = 4.13YY365 pKa = 10.38KK366 pKa = 10.11ICEE369 pKa = 4.15VNNPTNCDD377 pKa = 3.32SVISMIVVSAPIIAAVVDD395 pKa = 3.84TTAAVSGLTGGITSALTSNDD415 pKa = 3.39SLNGIVVVIGIDD427 pKa = 3.42SGNVTLTGLNVPAGLTLNADD447 pKa = 3.5GTVTVDD453 pKa = 3.18SNTPAGNYY461 pKa = 7.98NVEE464 pKa = 4.13YY465 pKa = 10.38KK466 pKa = 10.11ICEE469 pKa = 4.15VNNPTNCDD477 pKa = 3.32SVISMIVVSAPIIAAVVDD495 pKa = 3.84TTAAVSGLTGGITSALTSNDD515 pKa = 3.39SLNGIVVVIGIDD527 pKa = 3.24SGNVMLTGLTVPAGLTLNADD547 pKa = 3.5GTVTVDD553 pKa = 3.18SNTPAGNYY561 pKa = 7.98NVEE564 pKa = 4.13YY565 pKa = 10.38KK566 pKa = 10.11ICEE569 pKa = 4.15VNNPTNCDD577 pKa = 3.32SVISMIVVSAPIIAAVVDD595 pKa = 3.84TTAAVSGLTGGITSALTSNDD615 pKa = 3.39SLNGIVVVIGIDD627 pKa = 3.42SGNVTLTGLNVPAGLTLNADD647 pKa = 3.5GTVTVDD653 pKa = 3.18SNTPAGNYY661 pKa = 7.98NVEE664 pKa = 4.13YY665 pKa = 10.38KK666 pKa = 10.11ICEE669 pKa = 4.15VNNPTNCDD677 pKa = 3.3SVISMIEE684 pKa = 4.01VYY686 pKa = 10.36RR687 pKa = 11.84EE688 pKa = 4.03LPDD691 pKa = 3.88LSPTIDD697 pKa = 2.97IDD699 pKa = 4.03ALVFPIAGSAKK710 pKa = 10.29DD711 pKa = 3.52FVVNIGDD718 pKa = 3.8VKK720 pKa = 10.9GVEE723 pKa = 4.04SDD725 pKa = 3.52GQVVVKK731 pKa = 10.09VSKK734 pKa = 10.98GNAFFISYY742 pKa = 10.84ADD744 pKa = 3.45STSNSNANGGSVPVNNSAWIITEE767 pKa = 3.72NASFITITLKK777 pKa = 10.79PGVQISANTFSAIGFTITRR796 pKa = 11.84KK797 pKa = 10.31AGVPTQTSQPITVTIVNGTGLDD819 pKa = 3.75SQNSNNAYY827 pKa = 8.58STIVTAQQ834 pKa = 2.84
Molecular weight: 84.3 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z4GBV0|A0A2Z4GBV0_9BACT Glycosyltransferase family 2 protein OS=Arcticibacterium luteifluviistationis OX=1784714 GN=DJ013_10660 PE=4 SV=1
MM1 pKa = 7.47AVKK4 pKa = 10.11KK5 pKa = 10.09LKK7 pKa = 9.13PTTPGQRR14 pKa = 11.84QRR16 pKa = 11.84VAPDD20 pKa = 3.76FSDD23 pKa = 2.84ITTNKK28 pKa = 9.57PEE30 pKa = 3.92RR31 pKa = 11.84SLIGVVKK38 pKa = 9.62KK39 pKa = 9.32TGGRR43 pKa = 11.84NNEE46 pKa = 3.75GHH48 pKa = 5.56RR49 pKa = 11.84TSRR52 pKa = 11.84YY53 pKa = 8.83IGGGHH58 pKa = 5.9KK59 pKa = 9.68RR60 pKa = 11.84RR61 pKa = 11.84YY62 pKa = 9.75RR63 pKa = 11.84IIDD66 pKa = 3.74FKK68 pKa = 10.85RR69 pKa = 11.84DD70 pKa = 3.24KK71 pKa = 10.69RR72 pKa = 11.84DD73 pKa = 3.21MVAEE77 pKa = 4.23VATIEE82 pKa = 3.99YY83 pKa = 10.37DD84 pKa = 3.44PNRR87 pKa = 11.84SARR90 pKa = 11.84IALVKK95 pKa = 11.01YY96 pKa = 9.7EE97 pKa = 4.73DD98 pKa = 3.68GEE100 pKa = 4.03KK101 pKa = 10.23RR102 pKa = 11.84YY103 pKa = 10.2IIAPQGLQVGQKK115 pKa = 9.38IEE117 pKa = 4.6AGDD120 pKa = 3.89SVAPEE125 pKa = 3.88VGNCLRR131 pKa = 11.84VGNMPLGTIIHH142 pKa = 6.6NIEE145 pKa = 3.96LTPGKK150 pKa = 10.16GGQLSRR156 pKa = 11.84SAGTYY161 pKa = 9.86AQLLAKK167 pKa = 10.1DD168 pKa = 3.99GKK170 pKa = 10.57YY171 pKa = 8.43VTLKK175 pKa = 9.96MPSGEE180 pKa = 3.99MRR182 pKa = 11.84LVLSTCVATVGSVSNPDD199 pKa = 3.27HH200 pKa = 6.53MNVNLGKK207 pKa = 10.39AGRR210 pKa = 11.84NRR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84PRR219 pKa = 11.84VRR221 pKa = 11.84GVAMNPVDD229 pKa = 3.68HH230 pKa = 7.13PMGGGEE236 pKa = 3.89GRR238 pKa = 11.84ASGGHH243 pKa = 5.11PRR245 pKa = 11.84SRR247 pKa = 11.84NGVMAKK253 pKa = 10.11GKK255 pKa = 8.35KK256 pKa = 7.69TRR258 pKa = 11.84SKK260 pKa = 10.76NKK262 pKa = 9.01YY263 pKa = 7.5SNRR266 pKa = 11.84LIISKK271 pKa = 10.21RR272 pKa = 11.84SKK274 pKa = 10.67
MM1 pKa = 7.47AVKK4 pKa = 10.11KK5 pKa = 10.09LKK7 pKa = 9.13PTTPGQRR14 pKa = 11.84QRR16 pKa = 11.84VAPDD20 pKa = 3.76FSDD23 pKa = 2.84ITTNKK28 pKa = 9.57PEE30 pKa = 3.92RR31 pKa = 11.84SLIGVVKK38 pKa = 9.62KK39 pKa = 9.32TGGRR43 pKa = 11.84NNEE46 pKa = 3.75GHH48 pKa = 5.56RR49 pKa = 11.84TSRR52 pKa = 11.84YY53 pKa = 8.83IGGGHH58 pKa = 5.9KK59 pKa = 9.68RR60 pKa = 11.84RR61 pKa = 11.84YY62 pKa = 9.75RR63 pKa = 11.84IIDD66 pKa = 3.74FKK68 pKa = 10.85RR69 pKa = 11.84DD70 pKa = 3.24KK71 pKa = 10.69RR72 pKa = 11.84DD73 pKa = 3.21MVAEE77 pKa = 4.23VATIEE82 pKa = 3.99YY83 pKa = 10.37DD84 pKa = 3.44PNRR87 pKa = 11.84SARR90 pKa = 11.84IALVKK95 pKa = 11.01YY96 pKa = 9.7EE97 pKa = 4.73DD98 pKa = 3.68GEE100 pKa = 4.03KK101 pKa = 10.23RR102 pKa = 11.84YY103 pKa = 10.2IIAPQGLQVGQKK115 pKa = 9.38IEE117 pKa = 4.6AGDD120 pKa = 3.89SVAPEE125 pKa = 3.88VGNCLRR131 pKa = 11.84VGNMPLGTIIHH142 pKa = 6.6NIEE145 pKa = 3.96LTPGKK150 pKa = 10.16GGQLSRR156 pKa = 11.84SAGTYY161 pKa = 9.86AQLLAKK167 pKa = 10.1DD168 pKa = 3.99GKK170 pKa = 10.57YY171 pKa = 8.43VTLKK175 pKa = 9.96MPSGEE180 pKa = 3.99MRR182 pKa = 11.84LVLSTCVATVGSVSNPDD199 pKa = 3.27HH200 pKa = 6.53MNVNLGKK207 pKa = 10.39AGRR210 pKa = 11.84NRR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84PRR219 pKa = 11.84VRR221 pKa = 11.84GVAMNPVDD229 pKa = 3.68HH230 pKa = 7.13PMGGGEE236 pKa = 3.89GRR238 pKa = 11.84ASGGHH243 pKa = 5.11PRR245 pKa = 11.84SRR247 pKa = 11.84NGVMAKK253 pKa = 10.11GKK255 pKa = 8.35KK256 pKa = 7.69TRR258 pKa = 11.84SKK260 pKa = 10.76NKK262 pKa = 9.01YY263 pKa = 7.5SNRR266 pKa = 11.84LIISKK271 pKa = 10.21RR272 pKa = 11.84SKK274 pKa = 10.67
Molecular weight: 30.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1600995 |
38 |
7596 |
369.8 |
41.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.742 ± 0.04 | 0.808 ± 0.018 |
5.545 ± 0.031 | 6.59 ± 0.043 |
5.146 ± 0.039 | 7.099 ± 0.041 |
1.651 ± 0.017 | 7.31 ± 0.035 |
7.227 ± 0.055 | 9.372 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.218 ± 0.021 | 5.934 ± 0.047 |
3.594 ± 0.022 | 3.15 ± 0.023 |
3.391 ± 0.03 | 7.193 ± 0.056 |
5.756 ± 0.082 | 6.188 ± 0.029 |
1.178 ± 0.015 | 3.909 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
