
Beihai partiti-like virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.41
Get precalculated fractions of proteins
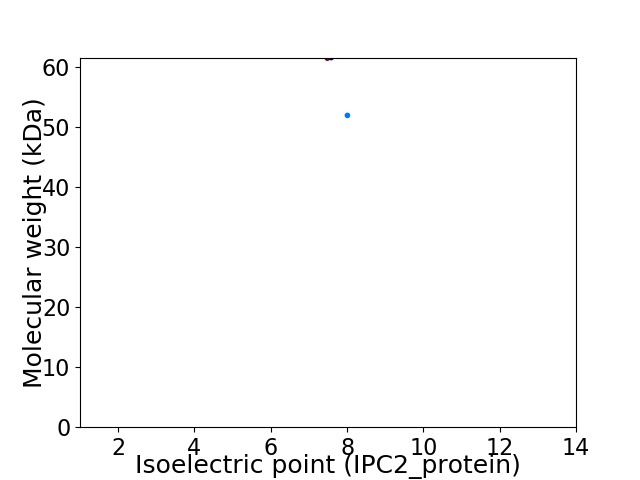
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KLI2|A0A1L3KLI2_9VIRU RdRp OS=Beihai partiti-like virus 2 OX=1922504 PE=4 SV=1
MM1 pKa = 7.25SLKK4 pKa = 10.3RR5 pKa = 11.84LPPSSSFPVDD15 pKa = 3.33TRR17 pKa = 11.84PPPVDD22 pKa = 3.74QIALKK27 pKa = 10.08YY28 pKa = 8.33ARR30 pKa = 11.84KK31 pKa = 8.89WFGDD35 pKa = 3.79GPVDD39 pKa = 3.46KK40 pKa = 11.07AIGSKK45 pKa = 10.01HH46 pKa = 5.81RR47 pKa = 11.84AVASNEE53 pKa = 3.92AIMDD57 pKa = 4.64NIRR60 pKa = 11.84GFDD63 pKa = 3.02RR64 pKa = 11.84RR65 pKa = 11.84YY66 pKa = 10.31APRR69 pKa = 11.84ITDD72 pKa = 3.92PVWEE76 pKa = 4.95SILQRR81 pKa = 11.84VYY83 pKa = 11.38EE84 pKa = 4.22EE85 pKa = 4.09IKK87 pKa = 10.48PNEE90 pKa = 4.27LIIPYY95 pKa = 7.06TTGGALKK102 pKa = 10.87DD103 pKa = 3.65PDD105 pKa = 4.16FPMKK109 pKa = 10.36KK110 pKa = 10.08SPGLPWKK117 pKa = 10.83LEE119 pKa = 4.22GYY121 pKa = 7.14GTKK124 pKa = 10.55GDD126 pKa = 3.4VCAIPEE132 pKa = 4.58NINKK136 pKa = 9.46IKK138 pKa = 10.34EE139 pKa = 3.77PWYY142 pKa = 9.52KK143 pKa = 10.33VGRR146 pKa = 11.84GYY148 pKa = 8.46HH149 pKa = 5.65TSMPDD154 pKa = 2.88CMVYY158 pKa = 10.23YY159 pKa = 10.41RR160 pKa = 11.84SHH162 pKa = 6.98ICDD165 pKa = 3.24TEE167 pKa = 4.12EE168 pKa = 3.76NKK170 pKa = 9.61IRR172 pKa = 11.84AVWGYY177 pKa = 8.22PLEE180 pKa = 4.44VFTEE184 pKa = 4.46EE185 pKa = 4.26ARR187 pKa = 11.84FVYY190 pKa = 10.07PFLQHH195 pKa = 7.1LKK197 pKa = 7.55TTPEE201 pKa = 4.82DD202 pKa = 3.59YY203 pKa = 10.72PIAYY207 pKa = 8.8GLEE210 pKa = 4.19MINGGMAYY218 pKa = 10.24LEE220 pKa = 4.62EE221 pKa = 4.65AWQQTRR227 pKa = 11.84AKK229 pKa = 9.46FAIMLDD235 pKa = 3.19WSSFDD240 pKa = 3.51QSIPPWLIRR249 pKa = 11.84DD250 pKa = 3.58AFSILEE256 pKa = 3.96RR257 pKa = 11.84CFDD260 pKa = 4.16FSHH263 pKa = 6.59VLASDD268 pKa = 3.79GKK270 pKa = 10.06IFPVDD275 pKa = 3.11PDD277 pKa = 3.2KK278 pKa = 10.83THH280 pKa = 6.81RR281 pKa = 11.84RR282 pKa = 11.84WKK284 pKa = 10.38RR285 pKa = 11.84LISYY289 pKa = 8.04FINTPFRR296 pKa = 11.84MPTGEE301 pKa = 4.47RR302 pKa = 11.84FLKK305 pKa = 10.83SGGVPSGSGFTNLIDD320 pKa = 4.24SIVNILVVRR329 pKa = 11.84YY330 pKa = 8.17ITYY333 pKa = 10.43HH334 pKa = 5.96NLGEE338 pKa = 4.56LPLHH342 pKa = 7.05DD343 pKa = 4.66MALGDD348 pKa = 4.33DD349 pKa = 3.9SVVYY353 pKa = 10.17TNGHH357 pKa = 6.39PSLEE361 pKa = 4.19RR362 pKa = 11.84MSEE365 pKa = 4.11LAQEE369 pKa = 4.27WFGMTINMRR378 pKa = 11.84KK379 pKa = 9.87SYY381 pKa = 7.92VTRR384 pKa = 11.84CARR387 pKa = 11.84NIQFLGYY394 pKa = 8.45YY395 pKa = 9.37NEE397 pKa = 4.21YY398 pKa = 10.78GMPFRR403 pKa = 11.84DD404 pKa = 3.78VQFLIASWIFPEE416 pKa = 5.33RR417 pKa = 11.84FQTPDD422 pKa = 3.21PAFTAIRR429 pKa = 11.84GLGQLWSTMNSTAAVFWHH447 pKa = 6.05YY448 pKa = 10.88AVSDD452 pKa = 4.18IISDD456 pKa = 3.98YY457 pKa = 10.97NLPSNWIEE465 pKa = 4.72DD466 pKa = 3.46IRR468 pKa = 11.84EE469 pKa = 4.03KK470 pKa = 10.94YY471 pKa = 10.25PNVLKK476 pKa = 10.55FLSLYY481 pKa = 10.64GITKK485 pKa = 10.13ISLPEE490 pKa = 3.94IDD492 pKa = 4.13ARR494 pKa = 11.84ASVFEE499 pKa = 3.93VTPRR503 pKa = 11.84IKK505 pKa = 10.35PLRR508 pKa = 11.84YY509 pKa = 8.9PKK511 pKa = 10.44RR512 pKa = 11.84RR513 pKa = 11.84DD514 pKa = 2.97LDD516 pKa = 3.25IRR518 pKa = 11.84SCYY521 pKa = 9.18QRR523 pKa = 11.84GRR525 pKa = 11.84DD526 pKa = 3.74VVPLRR531 pKa = 11.84DD532 pKa = 3.28
MM1 pKa = 7.25SLKK4 pKa = 10.3RR5 pKa = 11.84LPPSSSFPVDD15 pKa = 3.33TRR17 pKa = 11.84PPPVDD22 pKa = 3.74QIALKK27 pKa = 10.08YY28 pKa = 8.33ARR30 pKa = 11.84KK31 pKa = 8.89WFGDD35 pKa = 3.79GPVDD39 pKa = 3.46KK40 pKa = 11.07AIGSKK45 pKa = 10.01HH46 pKa = 5.81RR47 pKa = 11.84AVASNEE53 pKa = 3.92AIMDD57 pKa = 4.64NIRR60 pKa = 11.84GFDD63 pKa = 3.02RR64 pKa = 11.84RR65 pKa = 11.84YY66 pKa = 10.31APRR69 pKa = 11.84ITDD72 pKa = 3.92PVWEE76 pKa = 4.95SILQRR81 pKa = 11.84VYY83 pKa = 11.38EE84 pKa = 4.22EE85 pKa = 4.09IKK87 pKa = 10.48PNEE90 pKa = 4.27LIIPYY95 pKa = 7.06TTGGALKK102 pKa = 10.87DD103 pKa = 3.65PDD105 pKa = 4.16FPMKK109 pKa = 10.36KK110 pKa = 10.08SPGLPWKK117 pKa = 10.83LEE119 pKa = 4.22GYY121 pKa = 7.14GTKK124 pKa = 10.55GDD126 pKa = 3.4VCAIPEE132 pKa = 4.58NINKK136 pKa = 9.46IKK138 pKa = 10.34EE139 pKa = 3.77PWYY142 pKa = 9.52KK143 pKa = 10.33VGRR146 pKa = 11.84GYY148 pKa = 8.46HH149 pKa = 5.65TSMPDD154 pKa = 2.88CMVYY158 pKa = 10.23YY159 pKa = 10.41RR160 pKa = 11.84SHH162 pKa = 6.98ICDD165 pKa = 3.24TEE167 pKa = 4.12EE168 pKa = 3.76NKK170 pKa = 9.61IRR172 pKa = 11.84AVWGYY177 pKa = 8.22PLEE180 pKa = 4.44VFTEE184 pKa = 4.46EE185 pKa = 4.26ARR187 pKa = 11.84FVYY190 pKa = 10.07PFLQHH195 pKa = 7.1LKK197 pKa = 7.55TTPEE201 pKa = 4.82DD202 pKa = 3.59YY203 pKa = 10.72PIAYY207 pKa = 8.8GLEE210 pKa = 4.19MINGGMAYY218 pKa = 10.24LEE220 pKa = 4.62EE221 pKa = 4.65AWQQTRR227 pKa = 11.84AKK229 pKa = 9.46FAIMLDD235 pKa = 3.19WSSFDD240 pKa = 3.51QSIPPWLIRR249 pKa = 11.84DD250 pKa = 3.58AFSILEE256 pKa = 3.96RR257 pKa = 11.84CFDD260 pKa = 4.16FSHH263 pKa = 6.59VLASDD268 pKa = 3.79GKK270 pKa = 10.06IFPVDD275 pKa = 3.11PDD277 pKa = 3.2KK278 pKa = 10.83THH280 pKa = 6.81RR281 pKa = 11.84RR282 pKa = 11.84WKK284 pKa = 10.38RR285 pKa = 11.84LISYY289 pKa = 8.04FINTPFRR296 pKa = 11.84MPTGEE301 pKa = 4.47RR302 pKa = 11.84FLKK305 pKa = 10.83SGGVPSGSGFTNLIDD320 pKa = 4.24SIVNILVVRR329 pKa = 11.84YY330 pKa = 8.17ITYY333 pKa = 10.43HH334 pKa = 5.96NLGEE338 pKa = 4.56LPLHH342 pKa = 7.05DD343 pKa = 4.66MALGDD348 pKa = 4.33DD349 pKa = 3.9SVVYY353 pKa = 10.17TNGHH357 pKa = 6.39PSLEE361 pKa = 4.19RR362 pKa = 11.84MSEE365 pKa = 4.11LAQEE369 pKa = 4.27WFGMTINMRR378 pKa = 11.84KK379 pKa = 9.87SYY381 pKa = 7.92VTRR384 pKa = 11.84CARR387 pKa = 11.84NIQFLGYY394 pKa = 8.45YY395 pKa = 9.37NEE397 pKa = 4.21YY398 pKa = 10.78GMPFRR403 pKa = 11.84DD404 pKa = 3.78VQFLIASWIFPEE416 pKa = 5.33RR417 pKa = 11.84FQTPDD422 pKa = 3.21PAFTAIRR429 pKa = 11.84GLGQLWSTMNSTAAVFWHH447 pKa = 6.05YY448 pKa = 10.88AVSDD452 pKa = 4.18IISDD456 pKa = 3.98YY457 pKa = 10.97NLPSNWIEE465 pKa = 4.72DD466 pKa = 3.46IRR468 pKa = 11.84EE469 pKa = 4.03KK470 pKa = 10.94YY471 pKa = 10.25PNVLKK476 pKa = 10.55FLSLYY481 pKa = 10.64GITKK485 pKa = 10.13ISLPEE490 pKa = 3.94IDD492 pKa = 4.13ARR494 pKa = 11.84ASVFEE499 pKa = 3.93VTPRR503 pKa = 11.84IKK505 pKa = 10.35PLRR508 pKa = 11.84YY509 pKa = 8.9PKK511 pKa = 10.44RR512 pKa = 11.84RR513 pKa = 11.84DD514 pKa = 2.97LDD516 pKa = 3.25IRR518 pKa = 11.84SCYY521 pKa = 9.18QRR523 pKa = 11.84GRR525 pKa = 11.84DD526 pKa = 3.74VVPLRR531 pKa = 11.84DD532 pKa = 3.28
Molecular weight: 61.51 kDa
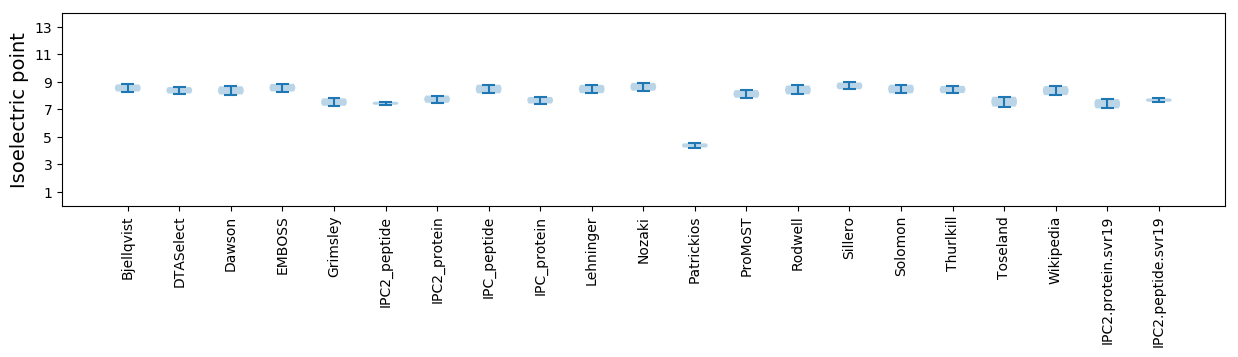
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KLI2|A0A1L3KLI2_9VIRU RdRp OS=Beihai partiti-like virus 2 OX=1922504 PE=4 SV=1
MM1 pKa = 7.57EE2 pKa = 6.22SIPEE6 pKa = 4.09NSNPAAPEE14 pKa = 4.14VPASAPRR21 pKa = 11.84GGGYY25 pKa = 9.74RR26 pKa = 11.84GRR28 pKa = 11.84GRR30 pKa = 11.84SGPRR34 pKa = 11.84GRR36 pKa = 11.84RR37 pKa = 11.84GRR39 pKa = 11.84GGGGRR44 pKa = 11.84GGQKK48 pKa = 9.79PQTPPPPPAPAGGDD62 pKa = 3.31PFAPRR67 pKa = 11.84GKK69 pKa = 9.94GGNPAIPKK77 pKa = 9.57DD78 pKa = 3.9DD79 pKa = 4.33PAADD83 pKa = 3.7SAIMIDD89 pKa = 4.38GIAGFVEE96 pKa = 4.31KK97 pKa = 10.08TKK99 pKa = 11.05NQLFQLDD106 pKa = 4.15LSQYY110 pKa = 7.81ITLVTEE116 pKa = 4.23SYY118 pKa = 10.37IAQTTCDD125 pKa = 3.11RR126 pKa = 11.84GLAKK130 pKa = 10.18FVSASAYY137 pKa = 8.71QYY139 pKa = 11.83YY140 pKa = 8.77MVILLWRR147 pKa = 11.84RR148 pKa = 11.84LFQLTSARR156 pKa = 11.84RR157 pKa = 11.84GDD159 pKa = 3.49SCFDD163 pKa = 3.48EE164 pKa = 5.24LSRR167 pKa = 11.84ALPHH171 pKa = 6.2SLPVPKK177 pKa = 10.06DD178 pKa = 2.96IKK180 pKa = 10.67IYY182 pKa = 10.66LDD184 pKa = 3.29SLGNIKK190 pKa = 10.61DD191 pKa = 3.82GEE193 pKa = 4.35GQDD196 pKa = 3.22WVFDD200 pKa = 5.07FIADD204 pKa = 4.32FSDD207 pKa = 3.19TKK209 pKa = 11.2VFGISGSYY217 pKa = 10.68GKK219 pKa = 10.26ISTEE223 pKa = 3.21NHH225 pKa = 5.15LQYY228 pKa = 10.2EE229 pKa = 4.56TVPCPVIPLLTMRR242 pKa = 11.84ADD244 pKa = 3.27ILRR247 pKa = 11.84TKK249 pKa = 10.58FAGTRR254 pKa = 11.84SWSLPTALTPADD266 pKa = 4.12VEE268 pKa = 4.45DD269 pKa = 4.59TSPGLPTLDD278 pKa = 3.32LLGYY282 pKa = 10.13QKK284 pKa = 11.22AKK286 pKa = 10.91NLTDD290 pKa = 3.36DD291 pKa = 3.88QYY293 pKa = 12.19NVLIGNGVDD302 pKa = 3.43VSGPAEE308 pKa = 4.47DD309 pKa = 3.38EE310 pKa = 4.49TFFNARR316 pKa = 11.84MVANVPVIQSLMGFLVGQFEE336 pKa = 4.66STKK339 pKa = 10.56CPMEE343 pKa = 5.55RR344 pKa = 11.84YY345 pKa = 7.72DD346 pKa = 3.46TTAITGSLAQVPFTVRR362 pKa = 11.84SSFSFSPNEE371 pKa = 4.17RR372 pKa = 11.84ISQKK376 pKa = 10.84VGITQTYY383 pKa = 7.93RR384 pKa = 11.84QSPPYY389 pKa = 8.86VACASMMFRR398 pKa = 11.84FRR400 pKa = 11.84IMRR403 pKa = 11.84AKK405 pKa = 10.45KK406 pKa = 10.38SGIQTDD412 pKa = 4.64GFCYY416 pKa = 9.9PWWNDD421 pKa = 3.0TKK423 pKa = 10.59KK424 pKa = 10.75KK425 pKa = 10.81YY426 pKa = 10.06IIPPEE431 pKa = 4.4WIANANDD438 pKa = 3.47IFNKK442 pKa = 8.54PEE444 pKa = 3.44KK445 pKa = 10.04WNISDD450 pKa = 4.2FTAGTADD457 pKa = 3.64GQALLSDD464 pKa = 4.78LARR467 pKa = 11.84KK468 pKa = 7.03TKK470 pKa = 10.21KK471 pKa = 10.05KK472 pKa = 10.59QEE474 pKa = 3.91
MM1 pKa = 7.57EE2 pKa = 6.22SIPEE6 pKa = 4.09NSNPAAPEE14 pKa = 4.14VPASAPRR21 pKa = 11.84GGGYY25 pKa = 9.74RR26 pKa = 11.84GRR28 pKa = 11.84GRR30 pKa = 11.84SGPRR34 pKa = 11.84GRR36 pKa = 11.84RR37 pKa = 11.84GRR39 pKa = 11.84GGGGRR44 pKa = 11.84GGQKK48 pKa = 9.79PQTPPPPPAPAGGDD62 pKa = 3.31PFAPRR67 pKa = 11.84GKK69 pKa = 9.94GGNPAIPKK77 pKa = 9.57DD78 pKa = 3.9DD79 pKa = 4.33PAADD83 pKa = 3.7SAIMIDD89 pKa = 4.38GIAGFVEE96 pKa = 4.31KK97 pKa = 10.08TKK99 pKa = 11.05NQLFQLDD106 pKa = 4.15LSQYY110 pKa = 7.81ITLVTEE116 pKa = 4.23SYY118 pKa = 10.37IAQTTCDD125 pKa = 3.11RR126 pKa = 11.84GLAKK130 pKa = 10.18FVSASAYY137 pKa = 8.71QYY139 pKa = 11.83YY140 pKa = 8.77MVILLWRR147 pKa = 11.84RR148 pKa = 11.84LFQLTSARR156 pKa = 11.84RR157 pKa = 11.84GDD159 pKa = 3.49SCFDD163 pKa = 3.48EE164 pKa = 5.24LSRR167 pKa = 11.84ALPHH171 pKa = 6.2SLPVPKK177 pKa = 10.06DD178 pKa = 2.96IKK180 pKa = 10.67IYY182 pKa = 10.66LDD184 pKa = 3.29SLGNIKK190 pKa = 10.61DD191 pKa = 3.82GEE193 pKa = 4.35GQDD196 pKa = 3.22WVFDD200 pKa = 5.07FIADD204 pKa = 4.32FSDD207 pKa = 3.19TKK209 pKa = 11.2VFGISGSYY217 pKa = 10.68GKK219 pKa = 10.26ISTEE223 pKa = 3.21NHH225 pKa = 5.15LQYY228 pKa = 10.2EE229 pKa = 4.56TVPCPVIPLLTMRR242 pKa = 11.84ADD244 pKa = 3.27ILRR247 pKa = 11.84TKK249 pKa = 10.58FAGTRR254 pKa = 11.84SWSLPTALTPADD266 pKa = 4.12VEE268 pKa = 4.45DD269 pKa = 4.59TSPGLPTLDD278 pKa = 3.32LLGYY282 pKa = 10.13QKK284 pKa = 11.22AKK286 pKa = 10.91NLTDD290 pKa = 3.36DD291 pKa = 3.88QYY293 pKa = 12.19NVLIGNGVDD302 pKa = 3.43VSGPAEE308 pKa = 4.47DD309 pKa = 3.38EE310 pKa = 4.49TFFNARR316 pKa = 11.84MVANVPVIQSLMGFLVGQFEE336 pKa = 4.66STKK339 pKa = 10.56CPMEE343 pKa = 5.55RR344 pKa = 11.84YY345 pKa = 7.72DD346 pKa = 3.46TTAITGSLAQVPFTVRR362 pKa = 11.84SSFSFSPNEE371 pKa = 4.17RR372 pKa = 11.84ISQKK376 pKa = 10.84VGITQTYY383 pKa = 7.93RR384 pKa = 11.84QSPPYY389 pKa = 8.86VACASMMFRR398 pKa = 11.84FRR400 pKa = 11.84IMRR403 pKa = 11.84AKK405 pKa = 10.45KK406 pKa = 10.38SGIQTDD412 pKa = 4.64GFCYY416 pKa = 9.9PWWNDD421 pKa = 3.0TKK423 pKa = 10.59KK424 pKa = 10.75KK425 pKa = 10.81YY426 pKa = 10.06IIPPEE431 pKa = 4.4WIANANDD438 pKa = 3.47IFNKK442 pKa = 8.54PEE444 pKa = 3.44KK445 pKa = 10.04WNISDD450 pKa = 4.2FTAGTADD457 pKa = 3.64GQALLSDD464 pKa = 4.78LARR467 pKa = 11.84KK468 pKa = 7.03TKK470 pKa = 10.21KK471 pKa = 10.05KK472 pKa = 10.59QEE474 pKa = 3.91
Molecular weight: 51.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1006 |
474 |
532 |
503.0 |
56.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.66 ± 0.9 | 1.193 ± 0.048 |
6.461 ± 0.052 | 4.672 ± 0.58 |
4.97 ± 0.078 | 7.455 ± 1.072 |
1.193 ± 0.511 | 6.859 ± 0.631 |
5.368 ± 0.218 | 7.058 ± 0.203 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.485 ± 0.249 | 3.579 ± 0.005 |
8.052 ± 0.117 | 3.28 ± 0.762 |
6.66 ± 0.499 | 6.958 ± 0.282 |
5.567 ± 0.645 | 5.07 ± 0.284 |
2.087 ± 0.405 | 4.374 ± 0.662 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
