
Jaminaea rosea
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Ustilaginomycotina; Exobasidiomycetes; Microstromatales; Microstromatales incertae sedis; Jaminaea
Average proteome isoelectric point is 6.86
Get precalculated fractions of proteins
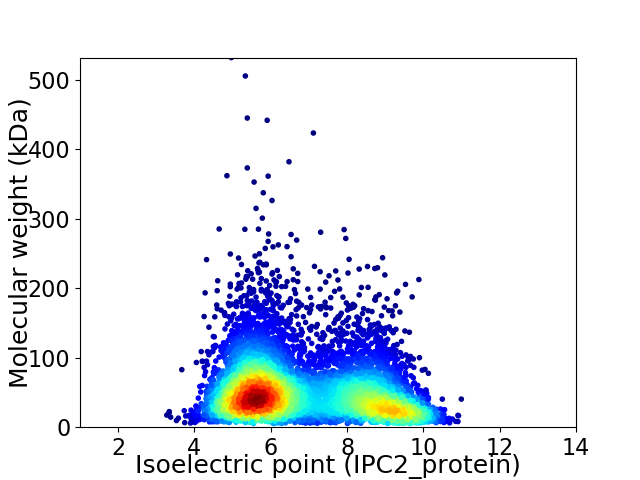
Virtual 2D-PAGE plot for 6858 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
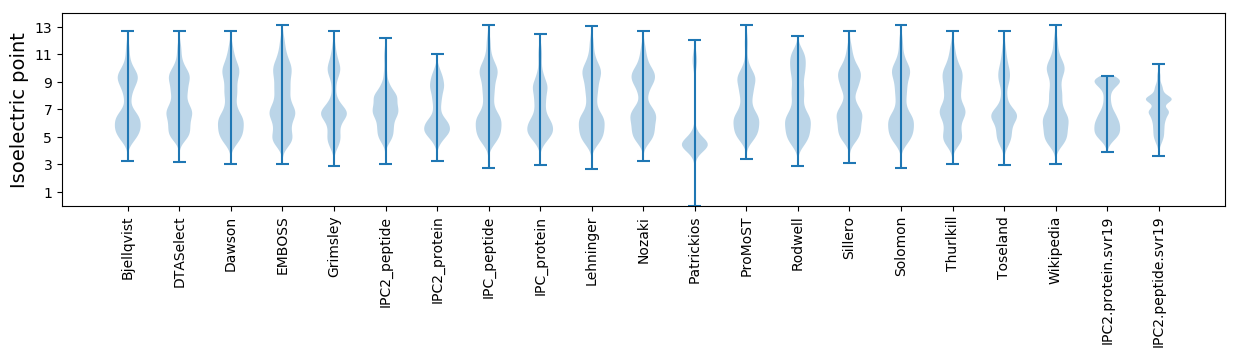
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A316UKN2|A0A316UKN2_9BASI DUF1162-domain-containing protein OS=Jaminaea rosea OX=1569628 GN=BDZ90DRAFT_267720 PE=3 SV=1
MM1 pKa = 7.33SPSFSSLFAFAALATTSLASSVTLQNSCSYY31 pKa = 9.28PVWVRR36 pKa = 11.84DD37 pKa = 3.7SSPGANGRR45 pKa = 11.84IVRR48 pKa = 11.84VNANSPSTDD57 pKa = 3.03VTQGGAGFVLNLLTGCSDD75 pKa = 4.25DD76 pKa = 4.79SGNGCATGDD85 pKa = 3.66VGGGALPFGRR95 pKa = 11.84VEE97 pKa = 4.04ATFTDD102 pKa = 4.03TYY104 pKa = 11.49VNYY107 pKa = 10.29DD108 pKa = 3.43LSLEE112 pKa = 4.31YY113 pKa = 10.57GWNVPVEE120 pKa = 4.47IGAPSCPYY128 pKa = 9.66FACTSLPGCPVPGPDD143 pKa = 3.61GSCYY147 pKa = 10.69SPCCASADD155 pKa = 3.65NCQGTNSCPYY165 pKa = 7.43EE166 pKa = 4.06TRR168 pKa = 11.84DD169 pKa = 3.63NGPGGKK175 pKa = 9.5GVNSQYY181 pKa = 11.91YY182 pKa = 9.4MDD184 pKa = 4.42TCPNAYY190 pKa = 9.48AWPIEE195 pKa = 4.39DD196 pKa = 3.74SDD198 pKa = 4.31VPSAYY203 pKa = 9.33IDD205 pKa = 4.04TNCGNQDD212 pKa = 3.5LQINFCPPVGNSNIPRR228 pKa = 11.84SS229 pKa = 3.65
MM1 pKa = 7.33SPSFSSLFAFAALATTSLASSVTLQNSCSYY31 pKa = 9.28PVWVRR36 pKa = 11.84DD37 pKa = 3.7SSPGANGRR45 pKa = 11.84IVRR48 pKa = 11.84VNANSPSTDD57 pKa = 3.03VTQGGAGFVLNLLTGCSDD75 pKa = 4.25DD76 pKa = 4.79SGNGCATGDD85 pKa = 3.66VGGGALPFGRR95 pKa = 11.84VEE97 pKa = 4.04ATFTDD102 pKa = 4.03TYY104 pKa = 11.49VNYY107 pKa = 10.29DD108 pKa = 3.43LSLEE112 pKa = 4.31YY113 pKa = 10.57GWNVPVEE120 pKa = 4.47IGAPSCPYY128 pKa = 9.66FACTSLPGCPVPGPDD143 pKa = 3.61GSCYY147 pKa = 10.69SPCCASADD155 pKa = 3.65NCQGTNSCPYY165 pKa = 7.43EE166 pKa = 4.06TRR168 pKa = 11.84DD169 pKa = 3.63NGPGGKK175 pKa = 9.5GVNSQYY181 pKa = 11.91YY182 pKa = 9.4MDD184 pKa = 4.42TCPNAYY190 pKa = 9.48AWPIEE195 pKa = 4.39DD196 pKa = 3.74SDD198 pKa = 4.31VPSAYY203 pKa = 9.33IDD205 pKa = 4.04TNCGNQDD212 pKa = 3.5LQINFCPPVGNSNIPRR228 pKa = 11.84SS229 pKa = 3.65
Molecular weight: 23.75 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A316US67|A0A316US67_9BASI Pseudouridine synthase OS=Jaminaea rosea OX=1569628 GN=BDZ90DRAFT_55210 PE=3 SV=1
MM1 pKa = 7.65NSALQDD7 pKa = 4.13AIKK10 pKa = 10.7NGQRR14 pKa = 11.84LKK16 pKa = 10.91KK17 pKa = 9.78ATTNDD22 pKa = 2.88RR23 pKa = 11.84SAPAVAGNAGGGGGGGGGGGGGAGGIAAAAAAARR57 pKa = 11.84TLASAPKK64 pKa = 10.21VPGTGGGSGGGGAAGLGGILAGGIPKK90 pKa = 10.53LKK92 pKa = 9.05PTGAAPAAPKK102 pKa = 10.1PPAASAPKK110 pKa = 9.86PPAAPGRR117 pKa = 11.84PVPPPTRR124 pKa = 11.84PSAPPAPPPPSRR136 pKa = 11.84PSAAAPPPPPPPGRR150 pKa = 11.84PAAPSAAAPPRR161 pKa = 11.84PPPPPPGRR169 pKa = 11.84SSPAVPPPPPPPARR183 pKa = 11.84PAAASPAAPAPPPPPPPGRR202 pKa = 11.84PAAAAPPPPSRR213 pKa = 11.84PSAAAPPPPPPPPAASSSAPARR235 pKa = 11.84GVPPPPSRR243 pKa = 11.84APAPPSRR250 pKa = 11.84PGAAPPAPPPPSTPARR266 pKa = 11.84GAVPPPPARR275 pKa = 11.84ASPISSAPPPPPSRR289 pKa = 11.84PASSAPPPPPPTRR302 pKa = 11.84SAAAAPAAPPPPPPSRR318 pKa = 11.84APAAAPPAPPPSRR331 pKa = 11.84APAAAPPTPPPSRR344 pKa = 11.84APAAPPPSRR353 pKa = 11.84SAAAAALPALNGAGSNGASRR373 pKa = 11.84SVSSTSISGGAAGKK387 pKa = 8.23TGRR390 pKa = 11.84IPSPPPTSGNLTFSPSSAFPPPRR413 pKa = 11.84AFAGSGASRR422 pKa = 11.84VYY424 pKa = 10.53PGGKK428 pKa = 7.54TRR430 pKa = 11.84GSTFDD435 pKa = 3.05WSTVGG440 pKa = 4.33
MM1 pKa = 7.65NSALQDD7 pKa = 4.13AIKK10 pKa = 10.7NGQRR14 pKa = 11.84LKK16 pKa = 10.91KK17 pKa = 9.78ATTNDD22 pKa = 2.88RR23 pKa = 11.84SAPAVAGNAGGGGGGGGGGGGGAGGIAAAAAAARR57 pKa = 11.84TLASAPKK64 pKa = 10.21VPGTGGGSGGGGAAGLGGILAGGIPKK90 pKa = 10.53LKK92 pKa = 9.05PTGAAPAAPKK102 pKa = 10.1PPAASAPKK110 pKa = 9.86PPAAPGRR117 pKa = 11.84PVPPPTRR124 pKa = 11.84PSAPPAPPPPSRR136 pKa = 11.84PSAAAPPPPPPPGRR150 pKa = 11.84PAAPSAAAPPRR161 pKa = 11.84PPPPPPGRR169 pKa = 11.84SSPAVPPPPPPPARR183 pKa = 11.84PAAASPAAPAPPPPPPPGRR202 pKa = 11.84PAAAAPPPPSRR213 pKa = 11.84PSAAAPPPPPPPPAASSSAPARR235 pKa = 11.84GVPPPPSRR243 pKa = 11.84APAPPSRR250 pKa = 11.84PGAAPPAPPPPSTPARR266 pKa = 11.84GAVPPPPARR275 pKa = 11.84ASPISSAPPPPPSRR289 pKa = 11.84PASSAPPPPPPTRR302 pKa = 11.84SAAAAPAAPPPPPPSRR318 pKa = 11.84APAAAPPAPPPSRR331 pKa = 11.84APAAAPPTPPPSRR344 pKa = 11.84APAAPPPSRR353 pKa = 11.84SAAAAALPALNGAGSNGASRR373 pKa = 11.84SVSSTSISGGAAGKK387 pKa = 8.23TGRR390 pKa = 11.84IPSPPPTSGNLTFSPSSAFPPPRR413 pKa = 11.84AFAGSGASRR422 pKa = 11.84VYY424 pKa = 10.53PGGKK428 pKa = 7.54TRR430 pKa = 11.84GSTFDD435 pKa = 3.05WSTVGG440 pKa = 4.33
Molecular weight: 40.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3700262 |
50 |
4870 |
539.6 |
58.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.072 ± 0.033 | 1.022 ± 0.011 |
5.454 ± 0.022 | 6.068 ± 0.026 |
2.918 ± 0.019 | 8.036 ± 0.032 |
2.358 ± 0.013 | 3.518 ± 0.021 |
4.175 ± 0.025 | 8.696 ± 0.032 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.897 ± 0.009 | 2.719 ± 0.013 |
6.42 ± 0.034 | 4.162 ± 0.018 |
6.923 ± 0.023 | 9.933 ± 0.048 |
5.588 ± 0.018 | 5.663 ± 0.02 |
1.292 ± 0.01 | 2.087 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
