
Acipenser ruthenus (Sterlet sturgeon)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Actinopterygii; Actinopteri; Chondrostei; Acipenseriformes; Acipenseroidei; Acipenseridae; Acipenserinae; Acipenserini; Acipenser
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
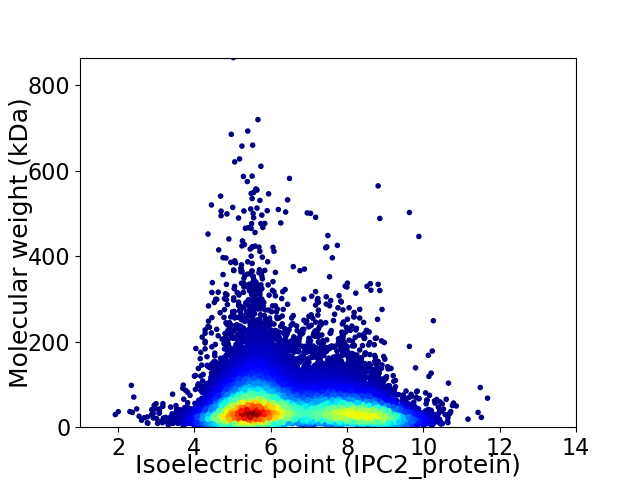
Virtual 2D-PAGE plot for 22004 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A444UUG6|A0A444UUG6_ACIRT NF-kappa-B inhibitor zeta OS=Acipenser ruthenus OX=7906 GN=EOD39_20792 PE=4 SV=1
MM1 pKa = 7.13YY2 pKa = 10.76VFDD5 pKa = 5.51IDD7 pKa = 4.86LKK9 pKa = 10.98SSTDD13 pKa = 3.54VAPLVCSDD21 pKa = 3.78VNEE24 pKa = 4.82CEE26 pKa = 4.16DD27 pKa = 3.89GRR29 pKa = 11.84NGGCVANSICSNTQGSFRR47 pKa = 11.84CGPCKK52 pKa = 10.1SGYY55 pKa = 10.12IGDD58 pKa = 3.72QRR60 pKa = 11.84QGCRR64 pKa = 11.84AKK66 pKa = 10.26QGCVNGQPSPCHH78 pKa = 6.62RR79 pKa = 11.84NAQCIPQRR87 pKa = 11.84DD88 pKa = 3.7GAVTCTCGVGWAGNGFMCGEE108 pKa = 4.4DD109 pKa = 3.35TDD111 pKa = 4.57IDD113 pKa = 3.96GFPDD117 pKa = 3.36EE118 pKa = 5.53KK119 pKa = 10.83LPCPEE124 pKa = 4.58TPCQKK129 pKa = 10.68DD130 pKa = 3.04NCLTVPNSGQEE141 pKa = 4.12DD142 pKa = 4.09ADD144 pKa = 3.68SDD146 pKa = 4.74GIGDD150 pKa = 4.47ACDD153 pKa = 3.53EE154 pKa = 4.74DD155 pKa = 5.69ADD157 pKa = 4.52GDD159 pKa = 4.51GILNMEE165 pKa = 4.99DD166 pKa = 3.17NCVLVPNVNQRR177 pKa = 11.84NVDD180 pKa = 3.13QDD182 pKa = 3.92VFGDD186 pKa = 3.45ACDD189 pKa = 3.48NCRR192 pKa = 11.84LINNNDD198 pKa = 3.19QRR200 pKa = 11.84DD201 pKa = 3.5TDD203 pKa = 4.0GDD205 pKa = 4.29SKK207 pKa = 11.76GDD209 pKa = 3.63ACDD212 pKa = 4.97DD213 pKa = 5.26DD214 pKa = 5.12IDD216 pKa = 5.99GDD218 pKa = 4.37GIRR221 pKa = 11.84NALDD225 pKa = 3.29NCEE228 pKa = 4.45KK229 pKa = 10.8VPNTDD234 pKa = 2.86QRR236 pKa = 11.84DD237 pKa = 3.52RR238 pKa = 11.84DD239 pKa = 3.6GDD241 pKa = 4.25GVGDD245 pKa = 4.36ACDD248 pKa = 3.45SCPDD252 pKa = 4.21DD253 pKa = 5.29RR254 pKa = 11.84NPDD257 pKa = 3.52QVGTTIYY264 pKa = 10.42KK265 pKa = 9.56HH266 pKa = 6.0PCVLYY271 pKa = 10.74HH272 pKa = 7.08LFNPWIIHH280 pKa = 6.99DD281 pKa = 4.26GDD283 pKa = 3.6GHH285 pKa = 6.51QDD287 pKa = 3.21SRR289 pKa = 11.84DD290 pKa = 3.54NCPTVINSSQLDD302 pKa = 3.55TDD304 pKa = 3.83RR305 pKa = 11.84DD306 pKa = 4.04GLGDD310 pKa = 3.86EE311 pKa = 5.62CDD313 pKa = 5.01DD314 pKa = 5.7DD315 pKa = 7.06DD316 pKa = 7.33DD317 pKa = 7.18GDD319 pKa = 4.76GIPDD323 pKa = 4.19EE324 pKa = 5.35LPPGPDD330 pKa = 3.02NCRR333 pKa = 11.84LIPNPRR339 pKa = 11.84QEE341 pKa = 5.43DD342 pKa = 3.67SDD344 pKa = 4.64GNNPWAA350 pKa = 4.68
MM1 pKa = 7.13YY2 pKa = 10.76VFDD5 pKa = 5.51IDD7 pKa = 4.86LKK9 pKa = 10.98SSTDD13 pKa = 3.54VAPLVCSDD21 pKa = 3.78VNEE24 pKa = 4.82CEE26 pKa = 4.16DD27 pKa = 3.89GRR29 pKa = 11.84NGGCVANSICSNTQGSFRR47 pKa = 11.84CGPCKK52 pKa = 10.1SGYY55 pKa = 10.12IGDD58 pKa = 3.72QRR60 pKa = 11.84QGCRR64 pKa = 11.84AKK66 pKa = 10.26QGCVNGQPSPCHH78 pKa = 6.62RR79 pKa = 11.84NAQCIPQRR87 pKa = 11.84DD88 pKa = 3.7GAVTCTCGVGWAGNGFMCGEE108 pKa = 4.4DD109 pKa = 3.35TDD111 pKa = 4.57IDD113 pKa = 3.96GFPDD117 pKa = 3.36EE118 pKa = 5.53KK119 pKa = 10.83LPCPEE124 pKa = 4.58TPCQKK129 pKa = 10.68DD130 pKa = 3.04NCLTVPNSGQEE141 pKa = 4.12DD142 pKa = 4.09ADD144 pKa = 3.68SDD146 pKa = 4.74GIGDD150 pKa = 4.47ACDD153 pKa = 3.53EE154 pKa = 4.74DD155 pKa = 5.69ADD157 pKa = 4.52GDD159 pKa = 4.51GILNMEE165 pKa = 4.99DD166 pKa = 3.17NCVLVPNVNQRR177 pKa = 11.84NVDD180 pKa = 3.13QDD182 pKa = 3.92VFGDD186 pKa = 3.45ACDD189 pKa = 3.48NCRR192 pKa = 11.84LINNNDD198 pKa = 3.19QRR200 pKa = 11.84DD201 pKa = 3.5TDD203 pKa = 4.0GDD205 pKa = 4.29SKK207 pKa = 11.76GDD209 pKa = 3.63ACDD212 pKa = 4.97DD213 pKa = 5.26DD214 pKa = 5.12IDD216 pKa = 5.99GDD218 pKa = 4.37GIRR221 pKa = 11.84NALDD225 pKa = 3.29NCEE228 pKa = 4.45KK229 pKa = 10.8VPNTDD234 pKa = 2.86QRR236 pKa = 11.84DD237 pKa = 3.52RR238 pKa = 11.84DD239 pKa = 3.6GDD241 pKa = 4.25GVGDD245 pKa = 4.36ACDD248 pKa = 3.45SCPDD252 pKa = 4.21DD253 pKa = 5.29RR254 pKa = 11.84NPDD257 pKa = 3.52QVGTTIYY264 pKa = 10.42KK265 pKa = 9.56HH266 pKa = 6.0PCVLYY271 pKa = 10.74HH272 pKa = 7.08LFNPWIIHH280 pKa = 6.99DD281 pKa = 4.26GDD283 pKa = 3.6GHH285 pKa = 6.51QDD287 pKa = 3.21SRR289 pKa = 11.84DD290 pKa = 3.54NCPTVINSSQLDD302 pKa = 3.55TDD304 pKa = 3.83RR305 pKa = 11.84DD306 pKa = 4.04GLGDD310 pKa = 3.86EE311 pKa = 5.62CDD313 pKa = 5.01DD314 pKa = 5.7DD315 pKa = 7.06DD316 pKa = 7.33DD317 pKa = 7.18GDD319 pKa = 4.76GIPDD323 pKa = 4.19EE324 pKa = 5.35LPPGPDD330 pKa = 3.02NCRR333 pKa = 11.84LIPNPRR339 pKa = 11.84QEE341 pKa = 5.43DD342 pKa = 3.67SDD344 pKa = 4.64GNNPWAA350 pKa = 4.68
Molecular weight: 37.58 kDa
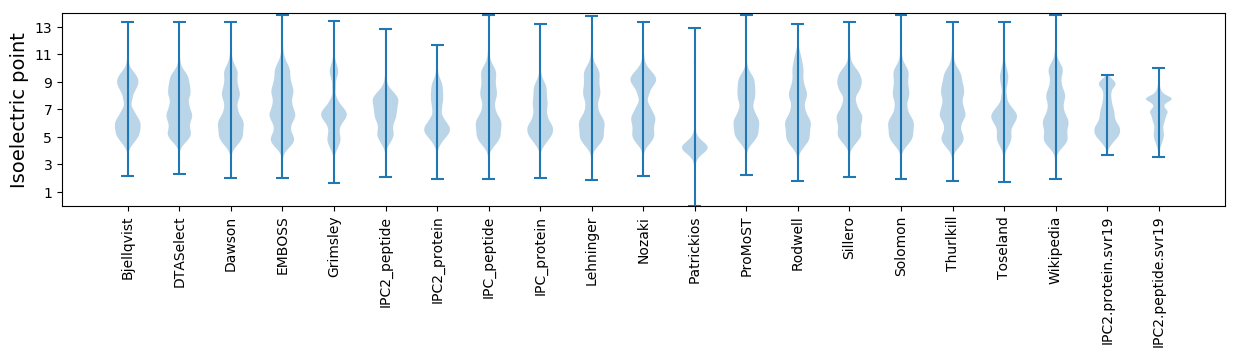
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A444USL8|A0A444USL8_ACIRT Leucine-rich repeat-containing protein 75A OS=Acipenser ruthenus OX=7906 GN=EOD39_21464 PE=4 SV=1
MM1 pKa = 7.08QRR3 pKa = 11.84NPMQSNPMQCNPMQRR18 pKa = 11.84NPMQSNPMQSNPMQHH33 pKa = 6.51NPMQRR38 pKa = 11.84NPMQHH43 pKa = 6.45NPMQRR48 pKa = 11.84NPMQRR53 pKa = 11.84NPMQRR58 pKa = 11.84NPMQRR63 pKa = 11.84NPMQRR68 pKa = 11.84NPMQRR73 pKa = 11.84NPMQRR78 pKa = 11.84NPMQRR83 pKa = 11.84NPMQRR88 pKa = 11.84NPMQRR93 pKa = 11.84NPMQRR98 pKa = 11.84NPMQRR103 pKa = 11.84NPMQRR108 pKa = 11.84NPMQRR113 pKa = 11.84NPMQRR118 pKa = 11.84NPMQRR123 pKa = 11.84NPMQRR128 pKa = 11.84NPMQRR133 pKa = 11.84NPMQRR138 pKa = 11.84NPMQRR143 pKa = 11.84NPMQRR148 pKa = 11.84NPMQRR153 pKa = 11.84NPMQRR158 pKa = 11.84NPMQRR163 pKa = 11.84NPMQRR168 pKa = 11.84NPMQRR173 pKa = 11.84NPMQRR178 pKa = 11.84NPMQRR183 pKa = 11.84NPMQRR188 pKa = 11.84NPMQRR193 pKa = 11.84NPMQRR198 pKa = 11.84NPMQRR203 pKa = 11.84NPMQRR208 pKa = 11.84NPMQRR213 pKa = 11.84NPMQRR218 pKa = 11.84NPMQRR223 pKa = 11.84NPMQRR228 pKa = 11.84NPMQRR233 pKa = 11.84NPMQHH238 pKa = 6.37NPMQHH243 pKa = 6.21NPMQHH248 pKa = 6.21NPMQHH253 pKa = 6.26NPMQRR258 pKa = 11.84NPMQHH263 pKa = 6.37NPMQHH268 pKa = 6.21NPMQHH273 pKa = 4.88NRR275 pKa = 11.84HH276 pKa = 5.47YY277 pKa = 10.7KK278 pKa = 9.39
MM1 pKa = 7.08QRR3 pKa = 11.84NPMQSNPMQCNPMQRR18 pKa = 11.84NPMQSNPMQSNPMQHH33 pKa = 6.51NPMQRR38 pKa = 11.84NPMQHH43 pKa = 6.45NPMQRR48 pKa = 11.84NPMQRR53 pKa = 11.84NPMQRR58 pKa = 11.84NPMQRR63 pKa = 11.84NPMQRR68 pKa = 11.84NPMQRR73 pKa = 11.84NPMQRR78 pKa = 11.84NPMQRR83 pKa = 11.84NPMQRR88 pKa = 11.84NPMQRR93 pKa = 11.84NPMQRR98 pKa = 11.84NPMQRR103 pKa = 11.84NPMQRR108 pKa = 11.84NPMQRR113 pKa = 11.84NPMQRR118 pKa = 11.84NPMQRR123 pKa = 11.84NPMQRR128 pKa = 11.84NPMQRR133 pKa = 11.84NPMQRR138 pKa = 11.84NPMQRR143 pKa = 11.84NPMQRR148 pKa = 11.84NPMQRR153 pKa = 11.84NPMQRR158 pKa = 11.84NPMQRR163 pKa = 11.84NPMQRR168 pKa = 11.84NPMQRR173 pKa = 11.84NPMQRR178 pKa = 11.84NPMQRR183 pKa = 11.84NPMQRR188 pKa = 11.84NPMQRR193 pKa = 11.84NPMQRR198 pKa = 11.84NPMQRR203 pKa = 11.84NPMQRR208 pKa = 11.84NPMQRR213 pKa = 11.84NPMQRR218 pKa = 11.84NPMQRR223 pKa = 11.84NPMQRR228 pKa = 11.84NPMQRR233 pKa = 11.84NPMQHH238 pKa = 6.37NPMQHH243 pKa = 6.21NPMQHH248 pKa = 6.21NPMQHH253 pKa = 6.26NPMQRR258 pKa = 11.84NPMQHH263 pKa = 6.37NPMQHH268 pKa = 6.21NPMQHH273 pKa = 4.88NRR275 pKa = 11.84HH276 pKa = 5.47YY277 pKa = 10.7KK278 pKa = 9.39
Molecular weight: 34.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
11239965 |
14 |
7624 |
510.8 |
57.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.459 ± 0.014 | 2.227 ± 0.016 |
5.091 ± 0.012 | 7.229 ± 0.025 |
3.586 ± 0.012 | 6.115 ± 0.02 |
2.575 ± 0.01 | 4.595 ± 0.015 |
6.071 ± 0.019 | 9.394 ± 0.022 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.347 ± 0.008 | 4.062 ± 0.012 |
5.502 ± 0.023 | 4.855 ± 0.018 |
5.367 ± 0.017 | 8.593 ± 0.024 |
5.616 ± 0.015 | 6.262 ± 0.015 |
1.115 ± 0.007 | 2.832 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
