
Pirellula staleyi (strain ATCC 27377 / DSM 6068 / ICPB 4128) (Pirella staleyi)
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Planctomycetia; Pirellulales; Pirellulaceae; Pirellula; Pirellula staleyi
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
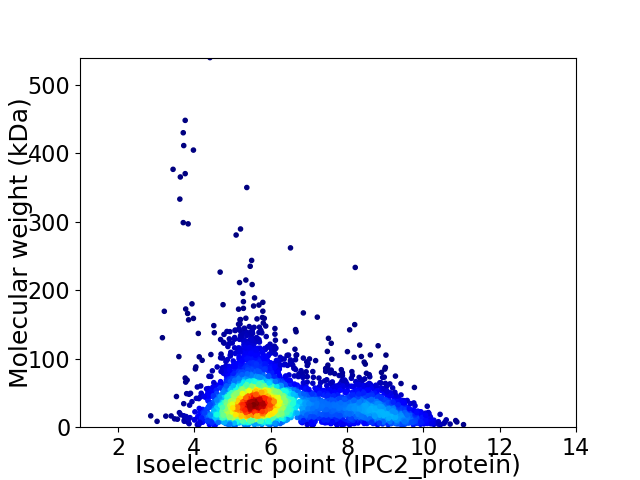
Virtual 2D-PAGE plot for 4711 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
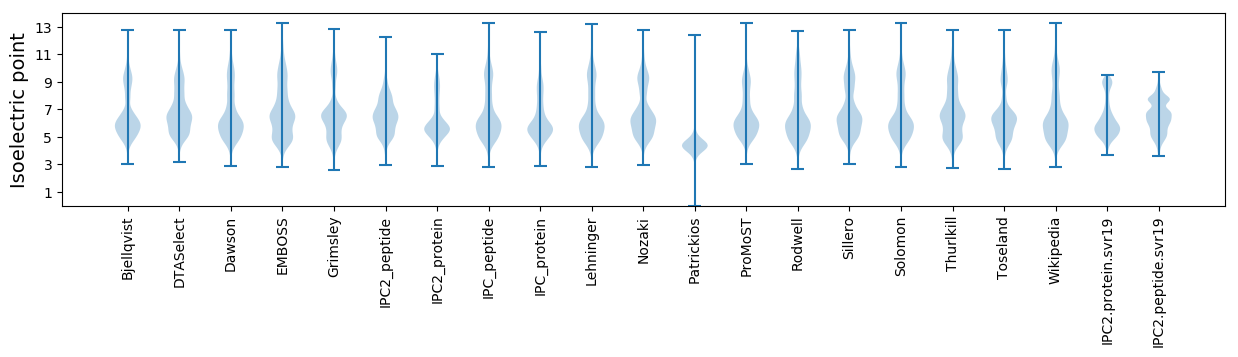
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D2R746|D2R746_PIRSD Uncharacterized protein OS=Pirellula staleyi (strain ATCC 27377 / DSM 6068 / ICPB 4128) OX=530564 GN=Psta_4607 PE=4 SV=1
MM1 pKa = 6.91KK2 pKa = 10.23HH3 pKa = 4.46LWKK6 pKa = 10.08WLGEE10 pKa = 3.91ALRR13 pKa = 11.84RR14 pKa = 11.84STGEE18 pKa = 3.68SSSRR22 pKa = 11.84SHH24 pKa = 6.47SRR26 pKa = 11.84SKK28 pKa = 10.66NRR30 pKa = 11.84RR31 pKa = 11.84GRR33 pKa = 11.84LRR35 pKa = 11.84LEE37 pKa = 4.13SLEE40 pKa = 4.03SRR42 pKa = 11.84AMMASDD48 pKa = 4.16ILPVLMVVADD58 pKa = 3.68QSDD61 pKa = 3.96FYY63 pKa = 9.91YY64 pKa = 10.54QEE66 pKa = 4.32YY67 pKa = 10.45GDD69 pKa = 3.47TRR71 pKa = 11.84MSIEE75 pKa = 3.88AAGLDD80 pKa = 3.68VQVAATTTNPTVPHH94 pKa = 6.16VNTGEE99 pKa = 4.16GADD102 pKa = 4.17GGLLTPDD109 pKa = 4.07LALADD114 pKa = 4.27VNASDD119 pKa = 3.8YY120 pKa = 11.09SAIVFVGGWGSSMYY134 pKa = 10.44QYY136 pKa = 11.08AFPGTYY142 pKa = 9.74SDD144 pKa = 3.31GRR146 pKa = 11.84YY147 pKa = 10.26NGDD150 pKa = 3.62LATKK154 pKa = 10.21GVVNEE159 pKa = 4.99LINDD163 pKa = 3.92FVEE166 pKa = 3.64QDD168 pKa = 3.22KK169 pKa = 11.24YY170 pKa = 9.63VTAICHH176 pKa = 5.38GTTVLAWARR185 pKa = 11.84VDD187 pKa = 4.17GVSLLAGRR195 pKa = 11.84TVAIPYY201 pKa = 9.37IGSPATFYY209 pKa = 10.29QGQYY213 pKa = 9.3YY214 pKa = 10.68GYY216 pKa = 10.69FEE218 pKa = 5.75LSQQAQMDD226 pKa = 3.98FNGAITQGSSGAIGDD241 pKa = 4.6PDD243 pKa = 3.71TAADD247 pKa = 4.19DD248 pKa = 3.63VWVDD252 pKa = 3.4GKK254 pKa = 10.88IITAEE259 pKa = 4.13NYY261 pKa = 10.28DD262 pKa = 3.69SAAMFGQVIASHH274 pKa = 6.81LLADD278 pKa = 5.05AEE280 pKa = 4.59GDD282 pKa = 4.04DD283 pKa = 4.42EE284 pKa = 5.49GGPIDD289 pKa = 5.02PPPPTNHH296 pKa = 6.88APTIADD302 pKa = 2.85GSMSIEE308 pKa = 4.17EE309 pKa = 4.07NSAIATLVVQVSGSDD324 pKa = 3.49VDD326 pKa = 4.72PEE328 pKa = 4.34DD329 pKa = 4.07LLTYY333 pKa = 10.46SIVSGNEE340 pKa = 3.4SGIFSIDD347 pKa = 3.07AATGEE352 pKa = 4.04IRR354 pKa = 11.84IASSTNLDD362 pKa = 3.69FEE364 pKa = 6.75ADD366 pKa = 3.51ALHH369 pKa = 6.01TLVVAVSDD377 pKa = 4.47GEE379 pKa = 4.3LSSEE383 pKa = 3.99ASIEE387 pKa = 4.11IAVLDD392 pKa = 3.91VDD394 pKa = 4.46EE395 pKa = 5.2TPPPPPMPLPPGPSNLTLPASPVAVVDD422 pKa = 4.07GVLIVQGTEE431 pKa = 3.85GADD434 pKa = 3.37YY435 pKa = 10.91LYY437 pKa = 10.65VYY439 pKa = 10.08PGAQAGSVMVYY450 pKa = 10.58RR451 pKa = 11.84NGQYY455 pKa = 10.67FSFAAGTVNSAVIHH469 pKa = 6.0ALGGDD474 pKa = 3.99DD475 pKa = 5.61FVGTCMLNLSVEE487 pKa = 4.26IYY489 pKa = 10.5GGNGHH494 pKa = 7.32DD495 pKa = 4.14EE496 pKa = 4.22LHH498 pKa = 6.33GAIAPSYY505 pKa = 9.7IDD507 pKa = 3.71GGEE510 pKa = 4.11GSDD513 pKa = 4.02RR514 pKa = 11.84IYY516 pKa = 11.29GGASSDD522 pKa = 3.71WIVGGNGDD530 pKa = 3.92DD531 pKa = 3.75QIEE534 pKa = 4.21ARR536 pKa = 11.84GGADD540 pKa = 3.5LVQGGAGTDD549 pKa = 3.81TINGGLGNDD558 pKa = 3.68ILIGGQGPDD567 pKa = 3.66RR568 pKa = 11.84LNGQADD574 pKa = 4.25DD575 pKa = 4.73DD576 pKa = 4.46LLIGGDD582 pKa = 3.82LQSAAVGASLLGLLAQWNTDD602 pKa = 2.84AATRR606 pKa = 11.84AQIWDD611 pKa = 3.52QGDD614 pKa = 3.41LGGDD618 pKa = 3.83FSVVGDD624 pKa = 4.12SAVDD628 pKa = 3.65CLFSGSGNDD637 pKa = 3.19WLLAGLTDD645 pKa = 4.34YY646 pKa = 10.66IYY648 pKa = 10.48QQEE651 pKa = 4.44PGDD654 pKa = 3.75EE655 pKa = 4.15LVRR658 pKa = 11.84RR659 pKa = 4.2
MM1 pKa = 6.91KK2 pKa = 10.23HH3 pKa = 4.46LWKK6 pKa = 10.08WLGEE10 pKa = 3.91ALRR13 pKa = 11.84RR14 pKa = 11.84STGEE18 pKa = 3.68SSSRR22 pKa = 11.84SHH24 pKa = 6.47SRR26 pKa = 11.84SKK28 pKa = 10.66NRR30 pKa = 11.84RR31 pKa = 11.84GRR33 pKa = 11.84LRR35 pKa = 11.84LEE37 pKa = 4.13SLEE40 pKa = 4.03SRR42 pKa = 11.84AMMASDD48 pKa = 4.16ILPVLMVVADD58 pKa = 3.68QSDD61 pKa = 3.96FYY63 pKa = 9.91YY64 pKa = 10.54QEE66 pKa = 4.32YY67 pKa = 10.45GDD69 pKa = 3.47TRR71 pKa = 11.84MSIEE75 pKa = 3.88AAGLDD80 pKa = 3.68VQVAATTTNPTVPHH94 pKa = 6.16VNTGEE99 pKa = 4.16GADD102 pKa = 4.17GGLLTPDD109 pKa = 4.07LALADD114 pKa = 4.27VNASDD119 pKa = 3.8YY120 pKa = 11.09SAIVFVGGWGSSMYY134 pKa = 10.44QYY136 pKa = 11.08AFPGTYY142 pKa = 9.74SDD144 pKa = 3.31GRR146 pKa = 11.84YY147 pKa = 10.26NGDD150 pKa = 3.62LATKK154 pKa = 10.21GVVNEE159 pKa = 4.99LINDD163 pKa = 3.92FVEE166 pKa = 3.64QDD168 pKa = 3.22KK169 pKa = 11.24YY170 pKa = 9.63VTAICHH176 pKa = 5.38GTTVLAWARR185 pKa = 11.84VDD187 pKa = 4.17GVSLLAGRR195 pKa = 11.84TVAIPYY201 pKa = 9.37IGSPATFYY209 pKa = 10.29QGQYY213 pKa = 9.3YY214 pKa = 10.68GYY216 pKa = 10.69FEE218 pKa = 5.75LSQQAQMDD226 pKa = 3.98FNGAITQGSSGAIGDD241 pKa = 4.6PDD243 pKa = 3.71TAADD247 pKa = 4.19DD248 pKa = 3.63VWVDD252 pKa = 3.4GKK254 pKa = 10.88IITAEE259 pKa = 4.13NYY261 pKa = 10.28DD262 pKa = 3.69SAAMFGQVIASHH274 pKa = 6.81LLADD278 pKa = 5.05AEE280 pKa = 4.59GDD282 pKa = 4.04DD283 pKa = 4.42EE284 pKa = 5.49GGPIDD289 pKa = 5.02PPPPTNHH296 pKa = 6.88APTIADD302 pKa = 2.85GSMSIEE308 pKa = 4.17EE309 pKa = 4.07NSAIATLVVQVSGSDD324 pKa = 3.49VDD326 pKa = 4.72PEE328 pKa = 4.34DD329 pKa = 4.07LLTYY333 pKa = 10.46SIVSGNEE340 pKa = 3.4SGIFSIDD347 pKa = 3.07AATGEE352 pKa = 4.04IRR354 pKa = 11.84IASSTNLDD362 pKa = 3.69FEE364 pKa = 6.75ADD366 pKa = 3.51ALHH369 pKa = 6.01TLVVAVSDD377 pKa = 4.47GEE379 pKa = 4.3LSSEE383 pKa = 3.99ASIEE387 pKa = 4.11IAVLDD392 pKa = 3.91VDD394 pKa = 4.46EE395 pKa = 5.2TPPPPPMPLPPGPSNLTLPASPVAVVDD422 pKa = 4.07GVLIVQGTEE431 pKa = 3.85GADD434 pKa = 3.37YY435 pKa = 10.91LYY437 pKa = 10.65VYY439 pKa = 10.08PGAQAGSVMVYY450 pKa = 10.58RR451 pKa = 11.84NGQYY455 pKa = 10.67FSFAAGTVNSAVIHH469 pKa = 6.0ALGGDD474 pKa = 3.99DD475 pKa = 5.61FVGTCMLNLSVEE487 pKa = 4.26IYY489 pKa = 10.5GGNGHH494 pKa = 7.32DD495 pKa = 4.14EE496 pKa = 4.22LHH498 pKa = 6.33GAIAPSYY505 pKa = 9.7IDD507 pKa = 3.71GGEE510 pKa = 4.11GSDD513 pKa = 4.02RR514 pKa = 11.84IYY516 pKa = 11.29GGASSDD522 pKa = 3.71WIVGGNGDD530 pKa = 3.92DD531 pKa = 3.75QIEE534 pKa = 4.21ARR536 pKa = 11.84GGADD540 pKa = 3.5LVQGGAGTDD549 pKa = 3.81TINGGLGNDD558 pKa = 3.68ILIGGQGPDD567 pKa = 3.66RR568 pKa = 11.84LNGQADD574 pKa = 4.25DD575 pKa = 4.73DD576 pKa = 4.46LLIGGDD582 pKa = 3.82LQSAAVGASLLGLLAQWNTDD602 pKa = 2.84AATRR606 pKa = 11.84AQIWDD611 pKa = 3.52QGDD614 pKa = 3.41LGGDD618 pKa = 3.83FSVVGDD624 pKa = 4.12SAVDD628 pKa = 3.65CLFSGSGNDD637 pKa = 3.19WLLAGLTDD645 pKa = 4.34YY646 pKa = 10.66IYY648 pKa = 10.48QQEE651 pKa = 4.44PGDD654 pKa = 3.75EE655 pKa = 4.15LVRR658 pKa = 11.84RR659 pKa = 4.2
Molecular weight: 68.83 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D2R1P8|D2R1P8_PIRSD Planctomycete cytochrome C OS=Pirellula staleyi (strain ATCC 27377 / DSM 6068 / ICPB 4128) OX=530564 GN=Psta_2093 PE=4 SV=1
MM1 pKa = 7.61AKK3 pKa = 9.85PHH5 pKa = 6.76RR6 pKa = 11.84SHH8 pKa = 7.16KK9 pKa = 9.8KK10 pKa = 8.31ANHH13 pKa = 5.91GKK15 pKa = 9.98RR16 pKa = 11.84PANAKK21 pKa = 9.59ARR23 pKa = 11.84RR24 pKa = 11.84AKK26 pKa = 9.74RR27 pKa = 11.84RR28 pKa = 11.84KK29 pKa = 9.54VRR31 pKa = 11.84TT32 pKa = 3.48
MM1 pKa = 7.61AKK3 pKa = 9.85PHH5 pKa = 6.76RR6 pKa = 11.84SHH8 pKa = 7.16KK9 pKa = 9.8KK10 pKa = 8.31ANHH13 pKa = 5.91GKK15 pKa = 9.98RR16 pKa = 11.84PANAKK21 pKa = 9.59ARR23 pKa = 11.84RR24 pKa = 11.84AKK26 pKa = 9.74RR27 pKa = 11.84RR28 pKa = 11.84KK29 pKa = 9.54VRR31 pKa = 11.84TT32 pKa = 3.48
Molecular weight: 3.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1765092 |
30 |
5010 |
374.7 |
40.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.629 ± 0.049 | 1.055 ± 0.015 |
5.441 ± 0.028 | 6.214 ± 0.045 |
3.449 ± 0.022 | 7.542 ± 0.056 |
2.062 ± 0.022 | 4.96 ± 0.024 |
4.013 ± 0.04 | 10.377 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.206 ± 0.02 | 2.841 ± 0.04 |
5.583 ± 0.04 | 3.96 ± 0.023 |
6.325 ± 0.044 | 6.681 ± 0.036 |
5.794 ± 0.048 | 7.079 ± 0.03 |
1.414 ± 0.014 | 2.374 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
