
Nocardioides euryhalodurans
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Propionibacteriales; Nocardioidaceae; Nocardioides
Average proteome isoelectric point is 5.87
Get precalculated fractions of proteins
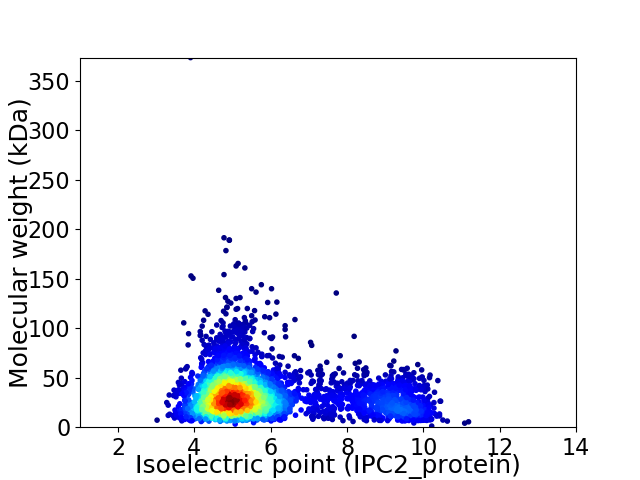
Virtual 2D-PAGE plot for 3773 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P7GLC7|A0A4P7GLC7_9ACTN Thioredoxin OS=Nocardioides euryhalodurans OX=2518370 GN=trxA PE=3 SV=1
MM1 pKa = 6.51TRR3 pKa = 11.84SRR5 pKa = 11.84ITSTACILASAALVTTGCAGPTPASEE31 pKa = 4.34GGEE34 pKa = 4.06WTPEE38 pKa = 3.43GDD40 pKa = 3.19MTMVVPFAAGGGSDD54 pKa = 3.37LSGRR58 pKa = 11.84AMAAGLEE65 pKa = 4.38GVQDD69 pKa = 3.99GLNVAVEE76 pKa = 4.18NRR78 pKa = 11.84DD79 pKa = 3.51GASGAVGYY87 pKa = 10.68SYY89 pKa = 11.35FLTKK93 pKa = 10.55EE94 pKa = 3.52GDD96 pKa = 3.56PNYY99 pKa = 10.79VLATEE104 pKa = 4.32TALLALPLTQDD115 pKa = 3.22VEE117 pKa = 4.21FDD119 pKa = 3.77YY120 pKa = 11.52TSFTPIMKK128 pKa = 10.04VGEE131 pKa = 4.61DD132 pKa = 3.5FTLLVVDD139 pKa = 4.83ADD141 pKa = 4.43SPYY144 pKa = 9.13EE145 pKa = 4.01TCTDD149 pKa = 3.24VVDD152 pKa = 3.73AAKK155 pKa = 10.25SEE157 pKa = 4.11RR158 pKa = 11.84VIAGISGEE166 pKa = 4.23TGLDD170 pKa = 3.06NVVFTLTEE178 pKa = 3.57QQMDD182 pKa = 3.84VEE184 pKa = 4.69FDD186 pKa = 3.39RR187 pKa = 11.84VPFEE191 pKa = 4.45SGSEE195 pKa = 3.81LTAALLGGTVDD206 pKa = 3.73IASLNPGEE214 pKa = 5.06VIGQLEE220 pKa = 4.25SGDD223 pKa = 3.61IKK225 pKa = 11.14ALCAYY230 pKa = 10.23ADD232 pKa = 3.44EE233 pKa = 5.65RR234 pKa = 11.84YY235 pKa = 10.25DD236 pKa = 3.78YY237 pKa = 10.87PEE239 pKa = 5.15LSDD242 pKa = 3.79IPTAKK247 pKa = 10.21EE248 pKa = 3.39QGIDD252 pKa = 3.31VSFAQFRR259 pKa = 11.84GLLATGGITEE269 pKa = 4.5SEE271 pKa = 3.96KK272 pKa = 10.88QGWIEE277 pKa = 3.76AAEE280 pKa = 4.16AFADD284 pKa = 3.4SDD286 pKa = 3.95AYY288 pKa = 9.79TEE290 pKa = 4.31YY291 pKa = 11.3VEE293 pKa = 5.33SNYY296 pKa = 9.95LQPVTAFGDD305 pKa = 3.79DD306 pKa = 3.47FTSYY310 pKa = 11.12LEE312 pKa = 4.52SNSEE316 pKa = 3.97TLAGVLEE323 pKa = 4.36
MM1 pKa = 6.51TRR3 pKa = 11.84SRR5 pKa = 11.84ITSTACILASAALVTTGCAGPTPASEE31 pKa = 4.34GGEE34 pKa = 4.06WTPEE38 pKa = 3.43GDD40 pKa = 3.19MTMVVPFAAGGGSDD54 pKa = 3.37LSGRR58 pKa = 11.84AMAAGLEE65 pKa = 4.38GVQDD69 pKa = 3.99GLNVAVEE76 pKa = 4.18NRR78 pKa = 11.84DD79 pKa = 3.51GASGAVGYY87 pKa = 10.68SYY89 pKa = 11.35FLTKK93 pKa = 10.55EE94 pKa = 3.52GDD96 pKa = 3.56PNYY99 pKa = 10.79VLATEE104 pKa = 4.32TALLALPLTQDD115 pKa = 3.22VEE117 pKa = 4.21FDD119 pKa = 3.77YY120 pKa = 11.52TSFTPIMKK128 pKa = 10.04VGEE131 pKa = 4.61DD132 pKa = 3.5FTLLVVDD139 pKa = 4.83ADD141 pKa = 4.43SPYY144 pKa = 9.13EE145 pKa = 4.01TCTDD149 pKa = 3.24VVDD152 pKa = 3.73AAKK155 pKa = 10.25SEE157 pKa = 4.11RR158 pKa = 11.84VIAGISGEE166 pKa = 4.23TGLDD170 pKa = 3.06NVVFTLTEE178 pKa = 3.57QQMDD182 pKa = 3.84VEE184 pKa = 4.69FDD186 pKa = 3.39RR187 pKa = 11.84VPFEE191 pKa = 4.45SGSEE195 pKa = 3.81LTAALLGGTVDD206 pKa = 3.73IASLNPGEE214 pKa = 5.06VIGQLEE220 pKa = 4.25SGDD223 pKa = 3.61IKK225 pKa = 11.14ALCAYY230 pKa = 10.23ADD232 pKa = 3.44EE233 pKa = 5.65RR234 pKa = 11.84YY235 pKa = 10.25DD236 pKa = 3.78YY237 pKa = 10.87PEE239 pKa = 5.15LSDD242 pKa = 3.79IPTAKK247 pKa = 10.21EE248 pKa = 3.39QGIDD252 pKa = 3.31VSFAQFRR259 pKa = 11.84GLLATGGITEE269 pKa = 4.5SEE271 pKa = 3.96KK272 pKa = 10.88QGWIEE277 pKa = 3.76AAEE280 pKa = 4.16AFADD284 pKa = 3.4SDD286 pKa = 3.95AYY288 pKa = 9.79TEE290 pKa = 4.31YY291 pKa = 11.3VEE293 pKa = 5.33SNYY296 pKa = 9.95LQPVTAFGDD305 pKa = 3.79DD306 pKa = 3.47FTSYY310 pKa = 11.12LEE312 pKa = 4.52SNSEE316 pKa = 3.97TLAGVLEE323 pKa = 4.36
Molecular weight: 33.98 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P7GKJ7|A0A4P7GKJ7_9ACTN Multidrug efflux SMR transporter OS=Nocardioides euryhalodurans OX=2518370 GN=EXE57_09890 PE=3 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.77HH17 pKa = 5.81RR18 pKa = 11.84KK19 pKa = 8.51LLKK22 pKa = 8.15KK23 pKa = 9.24TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 10.07LGKK33 pKa = 9.87
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.77HH17 pKa = 5.81RR18 pKa = 11.84KK19 pKa = 8.51LLKK22 pKa = 8.15KK23 pKa = 9.24TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 10.07LGKK33 pKa = 9.87
Molecular weight: 4.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1218950 |
11 |
3629 |
323.1 |
34.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.892 ± 0.056 | 0.72 ± 0.011 |
6.622 ± 0.037 | 6.07 ± 0.037 |
2.757 ± 0.022 | 9.39 ± 0.039 |
2.245 ± 0.022 | 3.12 ± 0.025 |
1.622 ± 0.026 | 10.519 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.831 ± 0.016 | 1.576 ± 0.021 |
5.632 ± 0.032 | 2.723 ± 0.019 |
7.971 ± 0.049 | 5.098 ± 0.023 |
6.054 ± 0.033 | 9.69 ± 0.037 |
1.569 ± 0.017 | 1.899 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
