
Chandiru virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Phenuiviridae; Phlebovirus; Candiru phlebovirus
Average proteome isoelectric point is 6.46
Get precalculated fractions of proteins
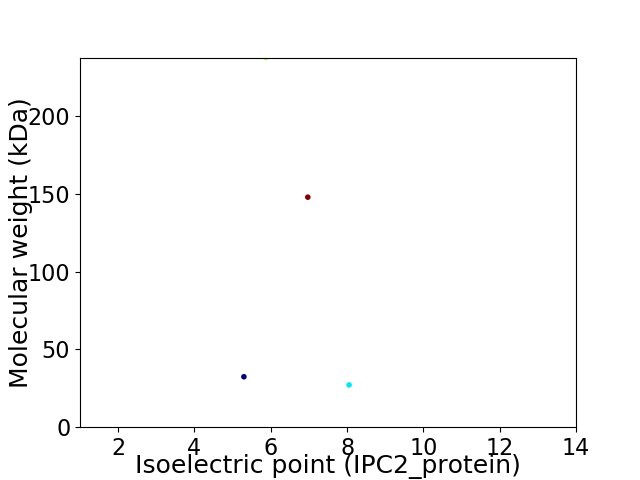
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
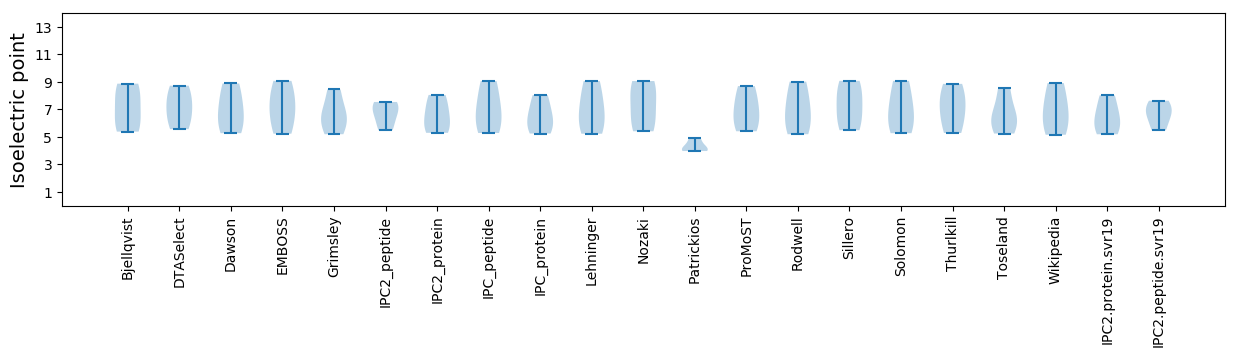
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F2W3Q3|F2W3Q3_9VIRU Nonstructural protein OS=Chandiru virus OX=629725 PE=4 SV=1
MM1 pKa = 7.93SNMLNNVFDD10 pKa = 3.97IPHH13 pKa = 6.19ITRR16 pKa = 11.84SATDD20 pKa = 3.22RR21 pKa = 11.84SKK23 pKa = 11.57VYY25 pKa = 10.74VEE27 pKa = 3.76YY28 pKa = 10.58SAYY31 pKa = 9.99NGYY34 pKa = 9.32WGTPICRR41 pKa = 11.84YY42 pKa = 9.78KK43 pKa = 10.84GAEE46 pKa = 4.29FPCSSFTVSDD56 pKa = 3.33KK57 pKa = 10.84TSFRR61 pKa = 11.84LRR63 pKa = 11.84SFIDD67 pKa = 3.03KK68 pKa = 10.62GQLPRR73 pKa = 11.84SWGKK77 pKa = 9.57PFLQVKK83 pKa = 9.59GKK85 pKa = 10.26SPSFFDD91 pKa = 3.31TTIEE95 pKa = 4.51KK96 pKa = 9.97ISHH99 pKa = 6.52LNLEE103 pKa = 4.68RR104 pKa = 11.84QLKK107 pKa = 8.9WSEE110 pKa = 4.12PNLKK114 pKa = 9.92EE115 pKa = 4.11ALSWPLGVPTLAFFKK130 pKa = 10.8LSLVEE135 pKa = 4.31SYY137 pKa = 10.81EE138 pKa = 4.2FNWEE142 pKa = 4.1EE143 pKa = 4.05KK144 pKa = 10.17CFLMTLIMRR153 pKa = 11.84SGDD156 pKa = 3.18GAQIDD161 pKa = 3.99DD162 pKa = 4.52SLVNLYY168 pKa = 10.46KK169 pKa = 10.75KK170 pKa = 10.69LIRR173 pKa = 11.84EE174 pKa = 4.05LGEE177 pKa = 4.03RR178 pKa = 11.84DD179 pKa = 3.46IPCLNFTGQNIEE191 pKa = 4.0KK192 pKa = 9.61EE193 pKa = 3.86IAYY196 pKa = 9.34VQVLRR201 pKa = 11.84MLTALPYY208 pKa = 10.31DD209 pKa = 4.01YY210 pKa = 11.08YY211 pKa = 11.1EE212 pKa = 5.13SDD214 pKa = 3.74FHH216 pKa = 8.58SSLYY220 pKa = 10.77DD221 pKa = 3.52YY222 pKa = 11.14VKK224 pKa = 10.47EE225 pKa = 3.83LSEE228 pKa = 4.7VITPHH233 pKa = 7.08LLGNRR238 pKa = 11.84KK239 pKa = 6.41WTPLHH244 pKa = 5.75TMSPRR249 pKa = 11.84YY250 pKa = 8.23TRR252 pKa = 11.84IADD255 pKa = 3.47SWDD258 pKa = 3.26SDD260 pKa = 3.74LDD262 pKa = 3.8SEE264 pKa = 5.02EE265 pKa = 5.14DD266 pKa = 3.63MEE268 pKa = 6.22IDD270 pKa = 3.88GPAARR275 pKa = 11.84WGEE278 pKa = 4.33GNN280 pKa = 3.61
MM1 pKa = 7.93SNMLNNVFDD10 pKa = 3.97IPHH13 pKa = 6.19ITRR16 pKa = 11.84SATDD20 pKa = 3.22RR21 pKa = 11.84SKK23 pKa = 11.57VYY25 pKa = 10.74VEE27 pKa = 3.76YY28 pKa = 10.58SAYY31 pKa = 9.99NGYY34 pKa = 9.32WGTPICRR41 pKa = 11.84YY42 pKa = 9.78KK43 pKa = 10.84GAEE46 pKa = 4.29FPCSSFTVSDD56 pKa = 3.33KK57 pKa = 10.84TSFRR61 pKa = 11.84LRR63 pKa = 11.84SFIDD67 pKa = 3.03KK68 pKa = 10.62GQLPRR73 pKa = 11.84SWGKK77 pKa = 9.57PFLQVKK83 pKa = 9.59GKK85 pKa = 10.26SPSFFDD91 pKa = 3.31TTIEE95 pKa = 4.51KK96 pKa = 9.97ISHH99 pKa = 6.52LNLEE103 pKa = 4.68RR104 pKa = 11.84QLKK107 pKa = 8.9WSEE110 pKa = 4.12PNLKK114 pKa = 9.92EE115 pKa = 4.11ALSWPLGVPTLAFFKK130 pKa = 10.8LSLVEE135 pKa = 4.31SYY137 pKa = 10.81EE138 pKa = 4.2FNWEE142 pKa = 4.1EE143 pKa = 4.05KK144 pKa = 10.17CFLMTLIMRR153 pKa = 11.84SGDD156 pKa = 3.18GAQIDD161 pKa = 3.99DD162 pKa = 4.52SLVNLYY168 pKa = 10.46KK169 pKa = 10.75KK170 pKa = 10.69LIRR173 pKa = 11.84EE174 pKa = 4.05LGEE177 pKa = 4.03RR178 pKa = 11.84DD179 pKa = 3.46IPCLNFTGQNIEE191 pKa = 4.0KK192 pKa = 9.61EE193 pKa = 3.86IAYY196 pKa = 9.34VQVLRR201 pKa = 11.84MLTALPYY208 pKa = 10.31DD209 pKa = 4.01YY210 pKa = 11.08YY211 pKa = 11.1EE212 pKa = 5.13SDD214 pKa = 3.74FHH216 pKa = 8.58SSLYY220 pKa = 10.77DD221 pKa = 3.52YY222 pKa = 11.14VKK224 pKa = 10.47EE225 pKa = 3.83LSEE228 pKa = 4.7VITPHH233 pKa = 7.08LLGNRR238 pKa = 11.84KK239 pKa = 6.41WTPLHH244 pKa = 5.75TMSPRR249 pKa = 11.84YY250 pKa = 8.23TRR252 pKa = 11.84IADD255 pKa = 3.47SWDD258 pKa = 3.26SDD260 pKa = 3.74LDD262 pKa = 3.8SEE264 pKa = 5.02EE265 pKa = 5.14DD266 pKa = 3.63MEE268 pKa = 6.22IDD270 pKa = 3.88GPAARR275 pKa = 11.84WGEE278 pKa = 4.33GNN280 pKa = 3.61
Molecular weight: 32.44 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F2W3Q3|F2W3Q3_9VIRU Nonstructural protein OS=Chandiru virus OX=629725 PE=4 SV=1
MM1 pKa = 8.05SYY3 pKa = 10.78EE4 pKa = 4.0KK5 pKa = 10.71LAVDD9 pKa = 4.1IAGHH13 pKa = 6.45EE14 pKa = 4.11IDD16 pKa = 5.31ADD18 pKa = 4.22TIKK21 pKa = 10.88AWVQAFAYY29 pKa = 10.12QGFDD33 pKa = 2.93AKK35 pKa = 10.46RR36 pKa = 11.84VMEE39 pKa = 4.34LLVEE43 pKa = 4.34RR44 pKa = 11.84GGDD47 pKa = 3.5DD48 pKa = 2.96WVEE51 pKa = 4.08DD52 pKa = 3.91AKK54 pKa = 11.56QMIILCLTRR63 pKa = 11.84GNKK66 pKa = 7.9PSKK69 pKa = 10.39MMVKK73 pKa = 9.95MSEE76 pKa = 3.84KK77 pKa = 10.19GKK79 pKa = 10.72KK80 pKa = 8.77IVQALVKK87 pKa = 10.1RR88 pKa = 11.84YY89 pKa = 8.3SLKK92 pKa = 10.6EE93 pKa = 4.01GNPSRR98 pKa = 11.84DD99 pKa = 3.73DD100 pKa = 3.48LTLSRR105 pKa = 11.84VTAALAGYY113 pKa = 6.71TCQATEE119 pKa = 4.08YY120 pKa = 10.23VEE122 pKa = 4.87EE123 pKa = 4.16FLPVTGKK130 pKa = 10.35NMDD133 pKa = 4.67DD134 pKa = 3.44LSKK137 pKa = 10.8NYY139 pKa = 8.62PRR141 pKa = 11.84AMMHH145 pKa = 6.7PSFAGLIDD153 pKa = 4.3PKK155 pKa = 10.81LPPDD159 pKa = 3.93VLSTICDD166 pKa = 3.36AFSLFMVQFSRR177 pKa = 11.84TINPRR182 pKa = 11.84NRR184 pKa = 11.84GLSVSEE190 pKa = 4.14VASTFDD196 pKa = 3.54RR197 pKa = 11.84PINAAMNSSFISGEE211 pKa = 3.72QRR213 pKa = 11.84KK214 pKa = 9.36SFLRR218 pKa = 11.84NLGILDD224 pKa = 4.11EE225 pKa = 4.86NMQPSNPVKK234 pKa = 10.34AAAKK238 pKa = 9.76VFRR241 pKa = 11.84GLKK244 pKa = 9.69
MM1 pKa = 8.05SYY3 pKa = 10.78EE4 pKa = 4.0KK5 pKa = 10.71LAVDD9 pKa = 4.1IAGHH13 pKa = 6.45EE14 pKa = 4.11IDD16 pKa = 5.31ADD18 pKa = 4.22TIKK21 pKa = 10.88AWVQAFAYY29 pKa = 10.12QGFDD33 pKa = 2.93AKK35 pKa = 10.46RR36 pKa = 11.84VMEE39 pKa = 4.34LLVEE43 pKa = 4.34RR44 pKa = 11.84GGDD47 pKa = 3.5DD48 pKa = 2.96WVEE51 pKa = 4.08DD52 pKa = 3.91AKK54 pKa = 11.56QMIILCLTRR63 pKa = 11.84GNKK66 pKa = 7.9PSKK69 pKa = 10.39MMVKK73 pKa = 9.95MSEE76 pKa = 3.84KK77 pKa = 10.19GKK79 pKa = 10.72KK80 pKa = 8.77IVQALVKK87 pKa = 10.1RR88 pKa = 11.84YY89 pKa = 8.3SLKK92 pKa = 10.6EE93 pKa = 4.01GNPSRR98 pKa = 11.84DD99 pKa = 3.73DD100 pKa = 3.48LTLSRR105 pKa = 11.84VTAALAGYY113 pKa = 6.71TCQATEE119 pKa = 4.08YY120 pKa = 10.23VEE122 pKa = 4.87EE123 pKa = 4.16FLPVTGKK130 pKa = 10.35NMDD133 pKa = 4.67DD134 pKa = 3.44LSKK137 pKa = 10.8NYY139 pKa = 8.62PRR141 pKa = 11.84AMMHH145 pKa = 6.7PSFAGLIDD153 pKa = 4.3PKK155 pKa = 10.81LPPDD159 pKa = 3.93VLSTICDD166 pKa = 3.36AFSLFMVQFSRR177 pKa = 11.84TINPRR182 pKa = 11.84NRR184 pKa = 11.84GLSVSEE190 pKa = 4.14VASTFDD196 pKa = 3.54RR197 pKa = 11.84PINAAMNSSFISGEE211 pKa = 3.72QRR213 pKa = 11.84KK214 pKa = 9.36SFLRR218 pKa = 11.84NLGILDD224 pKa = 4.11EE225 pKa = 4.86NMQPSNPVKK234 pKa = 10.34AAAKK238 pKa = 9.76VFRR241 pKa = 11.84GLKK244 pKa = 9.69
Molecular weight: 27.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3932 |
244 |
2088 |
983.0 |
111.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.705 ± 0.556 | 2.721 ± 0.762 |
5.722 ± 0.606 | 6.943 ± 0.268 |
4.73 ± 0.202 | 5.977 ± 0.421 |
2.314 ± 0.275 | 7.35 ± 0.586 |
7.248 ± 0.288 | 8.85 ± 0.208 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.077 ± 0.321 | 4.171 ± 0.399 |
3.662 ± 0.379 | 3.077 ± 0.245 |
4.959 ± 0.162 | 9.613 ± 0.255 |
4.807 ± 0.365 | 5.671 ± 0.191 |
1.348 ± 0.18 | 3.052 ± 0.238 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
