
Ginsengibacter hankyongi
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Chitinophagia; Chitinophagales; Chitinophagaceae; Ginsengibacter
Average proteome isoelectric point is 7.01
Get precalculated fractions of proteins
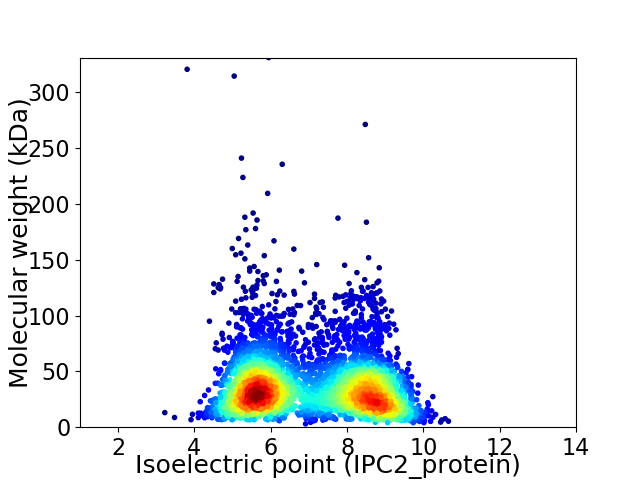
Virtual 2D-PAGE plot for 4476 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
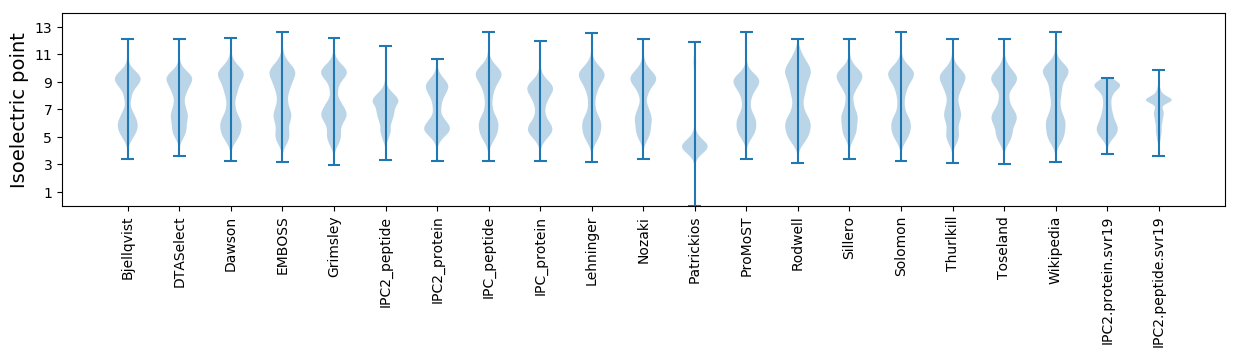
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5J5II15|A0A5J5II15_9BACT Uncharacterized protein OS=Ginsengibacter hankyongi OX=2607284 GN=FW778_10865 PE=4 SV=1
MM1 pKa = 7.55KK2 pKa = 10.51SKK4 pKa = 10.82LLLVVFTTLCFAANATTYY22 pKa = 10.6YY23 pKa = 10.19ISASGNDD30 pKa = 3.78ANNGTSTSTPWKK42 pKa = 10.26SLNKK46 pKa = 9.61LNSYY50 pKa = 10.44FSLLLPGDD58 pKa = 4.19NILFNRR64 pKa = 11.84GDD66 pKa = 3.41VFYY69 pKa = 11.58GNITVNASGKK79 pKa = 10.4AGLPLTISAYY89 pKa = 9.0GTGANPVITGFTNVTAWTNLGGNIWEE115 pKa = 4.41STNAISTLSTCNMVVINGVNTPMGRR140 pKa = 11.84WPNTGWLTYY149 pKa = 10.41EE150 pKa = 4.47SFSSSTSITSSSLNSSAINWTGATAVIKK178 pKa = 10.34KK179 pKa = 9.26EE180 pKa = 3.83RR181 pKa = 11.84YY182 pKa = 9.06IFTKK186 pKa = 9.03PTITSATGGTLNFASDD202 pKa = 3.58GHH204 pKa = 6.13NGRR207 pKa = 11.84ANWGFFIQNDD217 pKa = 3.54SRR219 pKa = 11.84TLDD222 pKa = 3.64MQNEE226 pKa = 4.56WYY228 pKa = 10.64YY229 pKa = 11.56NPSTKK234 pKa = 10.12KK235 pKa = 9.27IRR237 pKa = 11.84VYY239 pKa = 9.86STSIPVKK246 pKa = 8.65VQLAGVEE253 pKa = 4.16ALVTAINVSYY263 pKa = 10.64IAFDD267 pKa = 4.89GIDD270 pKa = 3.41FTGANTEE277 pKa = 3.78AFYY280 pKa = 11.33VGNFANCKK288 pKa = 9.85IINCSFNFNFQALQGKK304 pKa = 9.17ALGGSSANLLINNNTFNHH322 pKa = 6.13TNNNCIEE329 pKa = 4.29YY330 pKa = 8.58GTEE333 pKa = 4.01FTNSTVTNNTIKK345 pKa = 9.66NTSVFEE351 pKa = 4.02GMMGSGQSGWGISVTSSNTLIEE373 pKa = 4.31NNEE376 pKa = 3.95VDD378 pKa = 3.36TSGYY382 pKa = 9.41IAIGFNCTDD391 pKa = 3.26NSPNRR396 pKa = 11.84VKK398 pKa = 11.02NNYY401 pKa = 8.85VNYY404 pKa = 10.15FCFLKK409 pKa = 10.73DD410 pKa = 3.75DD411 pKa = 4.02GGGIYY416 pKa = 10.22SGNAQKK422 pKa = 10.82NQIIDD427 pKa = 3.33SNIIVNGIGNNTGTTSTNFSAVGIYY452 pKa = 10.52CDD454 pKa = 4.42DD455 pKa = 3.88NSSGFYY461 pKa = 10.63LRR463 pKa = 11.84NNTVANCNTGIYY475 pKa = 9.53LHH477 pKa = 6.53NANNIAVTYY486 pKa = 10.54NNIYY490 pKa = 10.74DD491 pKa = 3.99CGLGLFVSNDD501 pKa = 3.26NISFFTTNIYY511 pKa = 10.26SHH513 pKa = 6.54QNTIIAKK520 pKa = 5.86TTGADD525 pKa = 3.7FTPQDD530 pKa = 3.29QRR532 pKa = 11.84TVCFATIHH540 pKa = 5.82TSGNDD545 pKa = 2.67IRR547 pKa = 11.84NFGDD551 pKa = 3.02IDD553 pKa = 4.09SNYY556 pKa = 8.62YY557 pKa = 10.35ARR559 pKa = 11.84PIDD562 pKa = 4.2DD563 pKa = 4.39NLTIRR568 pKa = 11.84YY569 pKa = 6.08TLYY572 pKa = 10.89GAADD576 pKa = 3.58NDD578 pKa = 4.08ANLTMWQTYY587 pKa = 10.42SGFDD591 pKa = 3.47VHH593 pKa = 6.46SHH595 pKa = 6.88KK596 pKa = 10.9SPKK599 pKa = 10.0TITDD603 pKa = 3.75LNDD606 pKa = 3.42LLFSYY611 pKa = 10.87NSRR614 pKa = 11.84LVNNTSTLNATYY626 pKa = 10.21IDD628 pKa = 4.17VKK630 pKa = 10.75NISYY634 pKa = 9.78PGTITLPPYY643 pKa = 10.58SSAVLIKK650 pKa = 10.8NGATINQPPLANAGQDD666 pKa = 3.4QTITLPTNSLTLSGSGTDD684 pKa = 3.1SDD686 pKa = 4.44GTISSYY692 pKa = 9.84QWTKK696 pKa = 10.34ISGPSSYY703 pKa = 10.93SILNFTSPGTSITGLVQGIYY723 pKa = 10.17HH724 pKa = 6.32FQLTVTDD731 pKa = 3.99NNGATGIDD739 pKa = 3.62TMQVTVNPAPNQPPVTNAGSDD760 pKa = 3.47QTITLPLNAVTLTGSGSDD778 pKa = 3.47PDD780 pKa = 4.14GTISSYY786 pKa = 9.72QWTLISGPSGYY797 pKa = 10.82NIVNANSSGTDD808 pKa = 3.14VTGLVQGTYY817 pKa = 9.13QFQLTVTDD825 pKa = 4.07NNGATATDD833 pKa = 3.75VVLVTVNAALNISPIANAGSDD854 pKa = 3.51QTVTLPTNTITLSGSGSDD872 pKa = 3.17TDD874 pKa = 3.94GTVVSYY880 pKa = 10.45IWTEE884 pKa = 3.76VSGSSGYY891 pKa = 11.12NIVNANAPGTDD902 pKa = 3.15VTGLVQGTYY911 pKa = 9.13QFQLTVTDD919 pKa = 3.66NNGAIGTDD927 pKa = 2.91IMQVTVNSAPNISPVANAGTDD948 pKa = 3.38QTITLPANTLTLSGSGSDD966 pKa = 3.76ADD968 pKa = 3.71GTVVNYY974 pKa = 10.02AWTVISGPSSYY985 pKa = 11.22NIVNANSPGTDD996 pKa = 2.94VTGLVQGTYY1005 pKa = 9.07QFQLAVTDD1013 pKa = 3.75NNGAIGTDD1021 pKa = 3.68IIQVTVNAAPNQPPVANAGPDD1042 pKa = 3.39QTITLPTNTITLSGSGSDD1060 pKa = 3.32ADD1062 pKa = 3.97GTVVSYY1068 pKa = 10.64VWTVISGPSGYY1079 pKa = 10.83NIVNLNSPVTDD1090 pKa = 3.18VTGLLQGVYY1099 pKa = 10.06KK1100 pKa = 10.5FQLEE1104 pKa = 4.36VTDD1107 pKa = 4.41NNGATATDD1115 pKa = 3.35ITLVTVNAAPNISPVANAGMDD1136 pKa = 3.63QIITLPVDD1144 pKa = 4.14TITLSGSGSDD1154 pKa = 3.47ADD1156 pKa = 4.15GTISSYY1162 pKa = 9.67QWTEE1166 pKa = 3.03ISGPSSYY1173 pKa = 11.26NIVNANSPGTEE1184 pKa = 3.78VTGLVQGTYY1193 pKa = 9.13QFQLTVTDD1201 pKa = 3.66NNGAIGTDD1209 pKa = 3.09IMQLTANAAGNISPVANAGMDD1230 pKa = 3.63QIITLPVDD1238 pKa = 4.14TITLSGSGSDD1248 pKa = 3.32ADD1250 pKa = 3.97GTVVSYY1256 pKa = 11.02VWTAISGPSGYY1267 pKa = 10.78NIVNANSPGTEE1278 pKa = 3.78VTGLVQGTYY1287 pKa = 7.0QFRR1290 pKa = 11.84LTVTDD1295 pKa = 3.48NNGAIGTDD1303 pKa = 3.05IMQLTVNPAPNQPPVANAGTDD1324 pKa = 3.39QTITLPTNTVTLSGSGSDD1342 pKa = 3.15SDD1344 pKa = 4.11GTVVSYY1350 pKa = 8.96TWTEE1354 pKa = 3.3ISGPSSYY1361 pKa = 11.26NIVNANSPSTDD1372 pKa = 3.12VTGLAQGIYY1381 pKa = 9.69QFQLTVTDD1389 pKa = 4.63NIGATATDD1397 pKa = 3.36IVSITVNPVPNQPPVASAGIDD1418 pKa = 3.36QTITLPTNTVTLSGSGSDD1436 pKa = 3.15SDD1438 pKa = 4.1GTVVSYY1444 pKa = 10.66AWTMISGPSSYY1455 pKa = 11.15NIVNANSPSTDD1466 pKa = 2.86VTGLVQGVYY1475 pKa = 9.34QFQLTVIDD1483 pKa = 4.02NSGATATDD1491 pKa = 3.19IVSITVNAAANIPPVANAGIDD1512 pKa = 3.51QTITLPTNTVTLSGSGSDD1530 pKa = 3.15SDD1532 pKa = 4.11GTVVSYY1538 pKa = 8.96TWTEE1542 pKa = 3.3ISGPSGYY1549 pKa = 10.87NIVNPNSSFTDD1560 pKa = 3.22VTGLVEE1566 pKa = 4.87GVYY1569 pKa = 9.8QFQLQVTDD1577 pKa = 3.49NNGAIGTDD1585 pKa = 2.97IVQVTVNAVTNIPPVANAGLDD1606 pKa = 3.47QTITLPADD1614 pKa = 3.19SVTFSGIGSDD1624 pKa = 3.19ADD1626 pKa = 3.63GVVVSYY1632 pKa = 10.63AWSKK1636 pKa = 10.75ISGPSAYY1643 pKa = 9.57TIVNPALPSTNVTGLIQGVYY1663 pKa = 10.26LFEE1666 pKa = 5.01LQVTDD1671 pKa = 5.1DD1672 pKa = 4.25NGATSADD1679 pKa = 4.08TIQVTVNAATNIPPVANAGSDD1700 pKa = 3.48QTITLPTNSVTLFGNGSDD1718 pKa = 3.72ADD1720 pKa = 4.04GSIVGYY1726 pKa = 10.12NWTKK1730 pKa = 10.07ISGPSGYY1737 pKa = 9.08TIINATSASTDD1748 pKa = 3.13VTGLVQGLYY1757 pKa = 8.67QFRR1760 pKa = 11.84LKK1762 pKa = 9.95VTDD1765 pKa = 3.55NNGATGTDD1773 pKa = 4.09TIQVTVNISGNVSPVANAGSDD1794 pKa = 3.48QTITLPTDD1802 pKa = 3.15SVTLSGSGTDD1812 pKa = 3.17TDD1814 pKa = 4.13GTIVGYY1820 pKa = 8.2TWTQISGPSSYY1831 pKa = 11.13NILNATSSSTSVTGLVQGVYY1851 pKa = 9.31QFQLEE1856 pKa = 4.42VTDD1859 pKa = 4.07NSGATGTDD1867 pKa = 2.69IVRR1870 pKa = 11.84VIVNAPPNISPVANAGADD1888 pKa = 3.52QTIKK1892 pKa = 11.06LPVNSVTLSGSGTDD1906 pKa = 2.97SDD1908 pKa = 4.28GTVVSYY1914 pKa = 10.45EE1915 pKa = 3.72WTEE1918 pKa = 3.5ISGPSSYY1925 pKa = 11.15NIVNATSAATDD1936 pKa = 3.03ITGLVKK1942 pKa = 10.63GIYY1945 pKa = 9.77KK1946 pKa = 10.05FQLKK1950 pKa = 8.03VTDD1953 pKa = 3.7NDD1955 pKa = 4.0GATGTDD1961 pKa = 2.8IVKK1964 pKa = 8.8ITVNGSGNIVPVANAGSDD1982 pKa = 3.51QSITLPGNTITLSGSGTDD2000 pKa = 3.2ADD2002 pKa = 4.1GSVVSYY2008 pKa = 10.96NWTLMSGPSGYY2019 pKa = 10.92NIVNANLPATDD2030 pKa = 2.89ITGLVEE2036 pKa = 4.66GVYY2039 pKa = 9.82QFQLKK2044 pKa = 8.09VTDD2047 pKa = 3.56NKK2049 pKa = 10.93GAIGTDD2055 pKa = 2.86IVQVTVNSAPNQPPVANAGSDD2076 pKa = 3.62EE2077 pKa = 5.22SITLPTNSVTLSGSGTDD2094 pKa = 3.2ADD2096 pKa = 4.19GSVVSYY2102 pKa = 10.33KK2103 pKa = 8.45WTLISGPSGYY2113 pKa = 10.68SIINANLPATDD2124 pKa = 3.23VTGLVEE2130 pKa = 4.19GVYY2133 pKa = 9.92QFEE2136 pKa = 4.62LTVTDD2141 pKa = 3.37NSGAIGTDD2149 pKa = 3.55VIEE2152 pKa = 4.48VTVNAAPNTPPVANAGLDD2170 pKa = 3.67QIITLPTNTVKK2181 pKa = 10.62FSGSGSDD2188 pKa = 3.04VDD2190 pKa = 4.25GSVVSYY2196 pKa = 10.31KK2197 pKa = 8.45WTLISGPSGYY2207 pKa = 10.85NIVNANLPATDD2218 pKa = 3.23VTGLVEE2224 pKa = 5.04GIYY2227 pKa = 10.05QFEE2230 pKa = 4.54LTVTDD2235 pKa = 3.37NSGAIGTDD2243 pKa = 3.87VIQVTVNAMPNQLPVANAGPDD2264 pKa = 3.52QIITLPTNSLTLSGSGSDD2282 pKa = 3.46ADD2284 pKa = 4.06GTGVSYY2290 pKa = 10.79QWTKK2294 pKa = 10.44ISGPASYY2301 pKa = 10.86NILNGTLANSDD2312 pKa = 3.71VNNLVQGIYY2321 pKa = 9.73QFEE2324 pKa = 4.55LKK2326 pKa = 9.43VTDD2329 pKa = 3.88NNGATATDD2337 pKa = 3.23IMQVIVNAAPNQPPLANAGSDD2358 pKa = 3.89QIITLPTNSLTLSGSGSDD2376 pKa = 3.36ADD2378 pKa = 3.96GTIVSYY2384 pKa = 10.74QWTKK2388 pKa = 10.14ISGPASYY2395 pKa = 10.64NILNATPATVDD2406 pKa = 3.31VNNLVQGIYY2415 pKa = 9.73QFEE2418 pKa = 4.55LKK2420 pKa = 9.43VTDD2423 pKa = 3.88NNGATATDD2431 pKa = 3.45IMQLTVNAAPNQPPVANAGSDD2452 pKa = 3.7QIITLPTNSSTLSGSGSDD2470 pKa = 3.32ADD2472 pKa = 3.94GTVVSYY2478 pKa = 10.56KK2479 pKa = 6.35WTKK2482 pKa = 9.72ISGPSSYY2489 pKa = 10.49SIKK2492 pKa = 10.6DD2493 pKa = 3.27STSAITAINSLVEE2506 pKa = 3.75GVYY2509 pKa = 9.98QFEE2512 pKa = 4.64LTVTDD2517 pKa = 3.41NDD2519 pKa = 3.82GAIAIDD2525 pKa = 3.8EE2526 pKa = 4.42VQVTVNAAPNIPPVANAGTDD2546 pKa = 3.65KK2547 pKa = 10.93IITLPTNLVTISGSGNDD2564 pKa = 3.46ADD2566 pKa = 4.19GTIVSYY2572 pKa = 10.74QWTKK2576 pKa = 10.53ISGPANYY2583 pKa = 9.97NILDD2587 pKa = 3.92ATTATIDD2594 pKa = 3.38INSLVEE2600 pKa = 4.06GVYY2603 pKa = 9.85QFQLKK2608 pKa = 8.09VTDD2611 pKa = 3.86NKK2613 pKa = 10.58GASATDD2619 pKa = 3.2IMQVTVNAAPNVPPVANAGPDD2640 pKa = 3.51QIITLPANTLTISGSGSDD2658 pKa = 3.46ADD2660 pKa = 4.0GTVVGYY2666 pKa = 10.46LWTKK2670 pKa = 10.11VSGPANYY2677 pKa = 10.28NVVNPTSSVTGINGLVQGVYY2697 pKa = 9.42QFQLKK2702 pKa = 8.35ITDD2705 pKa = 3.66NKK2707 pKa = 10.48GATSTDD2713 pKa = 2.56IMQVIVNAAANKK2725 pKa = 10.41APVANAGADD2734 pKa = 3.55KK2735 pKa = 10.98SITLPTNSLTVSGSGSDD2752 pKa = 3.16ADD2754 pKa = 4.79GIVASYY2760 pKa = 9.67QWSKK2764 pKa = 11.02ISGPTSYY2771 pKa = 11.13NIVNPTSAVTDD2782 pKa = 3.4INGLVQGVYY2791 pKa = 8.92QFEE2794 pKa = 4.84LRR2796 pKa = 11.84VTDD2799 pKa = 3.53NNGARR2804 pKa = 11.84GRR2806 pKa = 11.84DD2807 pKa = 3.42TVQITVNAAVNIPPIAHH2824 pKa = 7.57AGPDD2828 pKa = 3.27ITITLPVNTASLAGSGSDD2846 pKa = 3.26ADD2848 pKa = 4.04GTVVSYY2854 pKa = 11.1VWTKK2858 pKa = 10.18ISGPSTYY2865 pKa = 10.76NIINASSPVTDD2876 pKa = 2.59VWGLLKK2882 pKa = 10.49GVYY2885 pKa = 9.6KK2886 pKa = 10.28FEE2888 pKa = 6.71IIVTDD2893 pKa = 3.55NKK2895 pKa = 10.84GATSRR2900 pKa = 11.84DD2901 pKa = 3.28TMQITVNSATNIAPVANAGKK2921 pKa = 9.57DD2922 pKa = 3.4QTITLPTNSVSLTGSGTDD2940 pKa = 3.11ADD2942 pKa = 4.23GTIVGYY2948 pKa = 7.91SWKK2951 pKa = 10.39QISGPSASVIASTNSAKK2968 pKa = 10.02TSANGLVGGTYY2979 pKa = 9.46EE2980 pKa = 4.98FEE2982 pKa = 4.14LTVIDD2987 pKa = 3.83NLGAVGKK2994 pKa = 8.56DD2995 pKa = 3.05TVAIVVAEE3003 pKa = 3.93PRR3005 pKa = 11.84LNLNVQTNTLMACPNPVIDD3024 pKa = 4.2EE3025 pKa = 4.31TTLEE3029 pKa = 4.12IKK3031 pKa = 9.88TSQVYY3036 pKa = 10.54SKK3038 pKa = 11.06LLLIVTDD3045 pKa = 3.82MNGKK3049 pKa = 8.96IVYY3052 pKa = 9.57KK3053 pKa = 10.72KK3054 pKa = 10.14EE3055 pKa = 3.54ITGGQTEE3062 pKa = 4.7LKK3064 pKa = 10.22EE3065 pKa = 4.03KK3066 pKa = 11.12LNMSNLIKK3074 pKa = 9.88GTYY3077 pKa = 9.97AITIYY3082 pKa = 11.02YY3083 pKa = 9.36SDD3085 pKa = 4.96KK3086 pKa = 9.65EE3087 pKa = 4.13KK3088 pKa = 11.05QSIKK3092 pKa = 10.49VIRR3095 pKa = 11.84LL3096 pKa = 3.24
MM1 pKa = 7.55KK2 pKa = 10.51SKK4 pKa = 10.82LLLVVFTTLCFAANATTYY22 pKa = 10.6YY23 pKa = 10.19ISASGNDD30 pKa = 3.78ANNGTSTSTPWKK42 pKa = 10.26SLNKK46 pKa = 9.61LNSYY50 pKa = 10.44FSLLLPGDD58 pKa = 4.19NILFNRR64 pKa = 11.84GDD66 pKa = 3.41VFYY69 pKa = 11.58GNITVNASGKK79 pKa = 10.4AGLPLTISAYY89 pKa = 9.0GTGANPVITGFTNVTAWTNLGGNIWEE115 pKa = 4.41STNAISTLSTCNMVVINGVNTPMGRR140 pKa = 11.84WPNTGWLTYY149 pKa = 10.41EE150 pKa = 4.47SFSSSTSITSSSLNSSAINWTGATAVIKK178 pKa = 10.34KK179 pKa = 9.26EE180 pKa = 3.83RR181 pKa = 11.84YY182 pKa = 9.06IFTKK186 pKa = 9.03PTITSATGGTLNFASDD202 pKa = 3.58GHH204 pKa = 6.13NGRR207 pKa = 11.84ANWGFFIQNDD217 pKa = 3.54SRR219 pKa = 11.84TLDD222 pKa = 3.64MQNEE226 pKa = 4.56WYY228 pKa = 10.64YY229 pKa = 11.56NPSTKK234 pKa = 10.12KK235 pKa = 9.27IRR237 pKa = 11.84VYY239 pKa = 9.86STSIPVKK246 pKa = 8.65VQLAGVEE253 pKa = 4.16ALVTAINVSYY263 pKa = 10.64IAFDD267 pKa = 4.89GIDD270 pKa = 3.41FTGANTEE277 pKa = 3.78AFYY280 pKa = 11.33VGNFANCKK288 pKa = 9.85IINCSFNFNFQALQGKK304 pKa = 9.17ALGGSSANLLINNNTFNHH322 pKa = 6.13TNNNCIEE329 pKa = 4.29YY330 pKa = 8.58GTEE333 pKa = 4.01FTNSTVTNNTIKK345 pKa = 9.66NTSVFEE351 pKa = 4.02GMMGSGQSGWGISVTSSNTLIEE373 pKa = 4.31NNEE376 pKa = 3.95VDD378 pKa = 3.36TSGYY382 pKa = 9.41IAIGFNCTDD391 pKa = 3.26NSPNRR396 pKa = 11.84VKK398 pKa = 11.02NNYY401 pKa = 8.85VNYY404 pKa = 10.15FCFLKK409 pKa = 10.73DD410 pKa = 3.75DD411 pKa = 4.02GGGIYY416 pKa = 10.22SGNAQKK422 pKa = 10.82NQIIDD427 pKa = 3.33SNIIVNGIGNNTGTTSTNFSAVGIYY452 pKa = 10.52CDD454 pKa = 4.42DD455 pKa = 3.88NSSGFYY461 pKa = 10.63LRR463 pKa = 11.84NNTVANCNTGIYY475 pKa = 9.53LHH477 pKa = 6.53NANNIAVTYY486 pKa = 10.54NNIYY490 pKa = 10.74DD491 pKa = 3.99CGLGLFVSNDD501 pKa = 3.26NISFFTTNIYY511 pKa = 10.26SHH513 pKa = 6.54QNTIIAKK520 pKa = 5.86TTGADD525 pKa = 3.7FTPQDD530 pKa = 3.29QRR532 pKa = 11.84TVCFATIHH540 pKa = 5.82TSGNDD545 pKa = 2.67IRR547 pKa = 11.84NFGDD551 pKa = 3.02IDD553 pKa = 4.09SNYY556 pKa = 8.62YY557 pKa = 10.35ARR559 pKa = 11.84PIDD562 pKa = 4.2DD563 pKa = 4.39NLTIRR568 pKa = 11.84YY569 pKa = 6.08TLYY572 pKa = 10.89GAADD576 pKa = 3.58NDD578 pKa = 4.08ANLTMWQTYY587 pKa = 10.42SGFDD591 pKa = 3.47VHH593 pKa = 6.46SHH595 pKa = 6.88KK596 pKa = 10.9SPKK599 pKa = 10.0TITDD603 pKa = 3.75LNDD606 pKa = 3.42LLFSYY611 pKa = 10.87NSRR614 pKa = 11.84LVNNTSTLNATYY626 pKa = 10.21IDD628 pKa = 4.17VKK630 pKa = 10.75NISYY634 pKa = 9.78PGTITLPPYY643 pKa = 10.58SSAVLIKK650 pKa = 10.8NGATINQPPLANAGQDD666 pKa = 3.4QTITLPTNSLTLSGSGTDD684 pKa = 3.1SDD686 pKa = 4.44GTISSYY692 pKa = 9.84QWTKK696 pKa = 10.34ISGPSSYY703 pKa = 10.93SILNFTSPGTSITGLVQGIYY723 pKa = 10.17HH724 pKa = 6.32FQLTVTDD731 pKa = 3.99NNGATGIDD739 pKa = 3.62TMQVTVNPAPNQPPVTNAGSDD760 pKa = 3.47QTITLPLNAVTLTGSGSDD778 pKa = 3.47PDD780 pKa = 4.14GTISSYY786 pKa = 9.72QWTLISGPSGYY797 pKa = 10.82NIVNANSSGTDD808 pKa = 3.14VTGLVQGTYY817 pKa = 9.13QFQLTVTDD825 pKa = 4.07NNGATATDD833 pKa = 3.75VVLVTVNAALNISPIANAGSDD854 pKa = 3.51QTVTLPTNTITLSGSGSDD872 pKa = 3.17TDD874 pKa = 3.94GTVVSYY880 pKa = 10.45IWTEE884 pKa = 3.76VSGSSGYY891 pKa = 11.12NIVNANAPGTDD902 pKa = 3.15VTGLVQGTYY911 pKa = 9.13QFQLTVTDD919 pKa = 3.66NNGAIGTDD927 pKa = 2.91IMQVTVNSAPNISPVANAGTDD948 pKa = 3.38QTITLPANTLTLSGSGSDD966 pKa = 3.76ADD968 pKa = 3.71GTVVNYY974 pKa = 10.02AWTVISGPSSYY985 pKa = 11.22NIVNANSPGTDD996 pKa = 2.94VTGLVQGTYY1005 pKa = 9.07QFQLAVTDD1013 pKa = 3.75NNGAIGTDD1021 pKa = 3.68IIQVTVNAAPNQPPVANAGPDD1042 pKa = 3.39QTITLPTNTITLSGSGSDD1060 pKa = 3.32ADD1062 pKa = 3.97GTVVSYY1068 pKa = 10.64VWTVISGPSGYY1079 pKa = 10.83NIVNLNSPVTDD1090 pKa = 3.18VTGLLQGVYY1099 pKa = 10.06KK1100 pKa = 10.5FQLEE1104 pKa = 4.36VTDD1107 pKa = 4.41NNGATATDD1115 pKa = 3.35ITLVTVNAAPNISPVANAGMDD1136 pKa = 3.63QIITLPVDD1144 pKa = 4.14TITLSGSGSDD1154 pKa = 3.47ADD1156 pKa = 4.15GTISSYY1162 pKa = 9.67QWTEE1166 pKa = 3.03ISGPSSYY1173 pKa = 11.26NIVNANSPGTEE1184 pKa = 3.78VTGLVQGTYY1193 pKa = 9.13QFQLTVTDD1201 pKa = 3.66NNGAIGTDD1209 pKa = 3.09IMQLTANAAGNISPVANAGMDD1230 pKa = 3.63QIITLPVDD1238 pKa = 4.14TITLSGSGSDD1248 pKa = 3.32ADD1250 pKa = 3.97GTVVSYY1256 pKa = 11.02VWTAISGPSGYY1267 pKa = 10.78NIVNANSPGTEE1278 pKa = 3.78VTGLVQGTYY1287 pKa = 7.0QFRR1290 pKa = 11.84LTVTDD1295 pKa = 3.48NNGAIGTDD1303 pKa = 3.05IMQLTVNPAPNQPPVANAGTDD1324 pKa = 3.39QTITLPTNTVTLSGSGSDD1342 pKa = 3.15SDD1344 pKa = 4.11GTVVSYY1350 pKa = 8.96TWTEE1354 pKa = 3.3ISGPSSYY1361 pKa = 11.26NIVNANSPSTDD1372 pKa = 3.12VTGLAQGIYY1381 pKa = 9.69QFQLTVTDD1389 pKa = 4.63NIGATATDD1397 pKa = 3.36IVSITVNPVPNQPPVASAGIDD1418 pKa = 3.36QTITLPTNTVTLSGSGSDD1436 pKa = 3.15SDD1438 pKa = 4.1GTVVSYY1444 pKa = 10.66AWTMISGPSSYY1455 pKa = 11.15NIVNANSPSTDD1466 pKa = 2.86VTGLVQGVYY1475 pKa = 9.34QFQLTVIDD1483 pKa = 4.02NSGATATDD1491 pKa = 3.19IVSITVNAAANIPPVANAGIDD1512 pKa = 3.51QTITLPTNTVTLSGSGSDD1530 pKa = 3.15SDD1532 pKa = 4.11GTVVSYY1538 pKa = 8.96TWTEE1542 pKa = 3.3ISGPSGYY1549 pKa = 10.87NIVNPNSSFTDD1560 pKa = 3.22VTGLVEE1566 pKa = 4.87GVYY1569 pKa = 9.8QFQLQVTDD1577 pKa = 3.49NNGAIGTDD1585 pKa = 2.97IVQVTVNAVTNIPPVANAGLDD1606 pKa = 3.47QTITLPADD1614 pKa = 3.19SVTFSGIGSDD1624 pKa = 3.19ADD1626 pKa = 3.63GVVVSYY1632 pKa = 10.63AWSKK1636 pKa = 10.75ISGPSAYY1643 pKa = 9.57TIVNPALPSTNVTGLIQGVYY1663 pKa = 10.26LFEE1666 pKa = 5.01LQVTDD1671 pKa = 5.1DD1672 pKa = 4.25NGATSADD1679 pKa = 4.08TIQVTVNAATNIPPVANAGSDD1700 pKa = 3.48QTITLPTNSVTLFGNGSDD1718 pKa = 3.72ADD1720 pKa = 4.04GSIVGYY1726 pKa = 10.12NWTKK1730 pKa = 10.07ISGPSGYY1737 pKa = 9.08TIINATSASTDD1748 pKa = 3.13VTGLVQGLYY1757 pKa = 8.67QFRR1760 pKa = 11.84LKK1762 pKa = 9.95VTDD1765 pKa = 3.55NNGATGTDD1773 pKa = 4.09TIQVTVNISGNVSPVANAGSDD1794 pKa = 3.48QTITLPTDD1802 pKa = 3.15SVTLSGSGTDD1812 pKa = 3.17TDD1814 pKa = 4.13GTIVGYY1820 pKa = 8.2TWTQISGPSSYY1831 pKa = 11.13NILNATSSSTSVTGLVQGVYY1851 pKa = 9.31QFQLEE1856 pKa = 4.42VTDD1859 pKa = 4.07NSGATGTDD1867 pKa = 2.69IVRR1870 pKa = 11.84VIVNAPPNISPVANAGADD1888 pKa = 3.52QTIKK1892 pKa = 11.06LPVNSVTLSGSGTDD1906 pKa = 2.97SDD1908 pKa = 4.28GTVVSYY1914 pKa = 10.45EE1915 pKa = 3.72WTEE1918 pKa = 3.5ISGPSSYY1925 pKa = 11.15NIVNATSAATDD1936 pKa = 3.03ITGLVKK1942 pKa = 10.63GIYY1945 pKa = 9.77KK1946 pKa = 10.05FQLKK1950 pKa = 8.03VTDD1953 pKa = 3.7NDD1955 pKa = 4.0GATGTDD1961 pKa = 2.8IVKK1964 pKa = 8.8ITVNGSGNIVPVANAGSDD1982 pKa = 3.51QSITLPGNTITLSGSGTDD2000 pKa = 3.2ADD2002 pKa = 4.1GSVVSYY2008 pKa = 10.96NWTLMSGPSGYY2019 pKa = 10.92NIVNANLPATDD2030 pKa = 2.89ITGLVEE2036 pKa = 4.66GVYY2039 pKa = 9.82QFQLKK2044 pKa = 8.09VTDD2047 pKa = 3.56NKK2049 pKa = 10.93GAIGTDD2055 pKa = 2.86IVQVTVNSAPNQPPVANAGSDD2076 pKa = 3.62EE2077 pKa = 5.22SITLPTNSVTLSGSGTDD2094 pKa = 3.2ADD2096 pKa = 4.19GSVVSYY2102 pKa = 10.33KK2103 pKa = 8.45WTLISGPSGYY2113 pKa = 10.68SIINANLPATDD2124 pKa = 3.23VTGLVEE2130 pKa = 4.19GVYY2133 pKa = 9.92QFEE2136 pKa = 4.62LTVTDD2141 pKa = 3.37NSGAIGTDD2149 pKa = 3.55VIEE2152 pKa = 4.48VTVNAAPNTPPVANAGLDD2170 pKa = 3.67QIITLPTNTVKK2181 pKa = 10.62FSGSGSDD2188 pKa = 3.04VDD2190 pKa = 4.25GSVVSYY2196 pKa = 10.31KK2197 pKa = 8.45WTLISGPSGYY2207 pKa = 10.85NIVNANLPATDD2218 pKa = 3.23VTGLVEE2224 pKa = 5.04GIYY2227 pKa = 10.05QFEE2230 pKa = 4.54LTVTDD2235 pKa = 3.37NSGAIGTDD2243 pKa = 3.87VIQVTVNAMPNQLPVANAGPDD2264 pKa = 3.52QIITLPTNSLTLSGSGSDD2282 pKa = 3.46ADD2284 pKa = 4.06GTGVSYY2290 pKa = 10.79QWTKK2294 pKa = 10.44ISGPASYY2301 pKa = 10.86NILNGTLANSDD2312 pKa = 3.71VNNLVQGIYY2321 pKa = 9.73QFEE2324 pKa = 4.55LKK2326 pKa = 9.43VTDD2329 pKa = 3.88NNGATATDD2337 pKa = 3.23IMQVIVNAAPNQPPLANAGSDD2358 pKa = 3.89QIITLPTNSLTLSGSGSDD2376 pKa = 3.36ADD2378 pKa = 3.96GTIVSYY2384 pKa = 10.74QWTKK2388 pKa = 10.14ISGPASYY2395 pKa = 10.64NILNATPATVDD2406 pKa = 3.31VNNLVQGIYY2415 pKa = 9.73QFEE2418 pKa = 4.55LKK2420 pKa = 9.43VTDD2423 pKa = 3.88NNGATATDD2431 pKa = 3.45IMQLTVNAAPNQPPVANAGSDD2452 pKa = 3.7QIITLPTNSSTLSGSGSDD2470 pKa = 3.32ADD2472 pKa = 3.94GTVVSYY2478 pKa = 10.56KK2479 pKa = 6.35WTKK2482 pKa = 9.72ISGPSSYY2489 pKa = 10.49SIKK2492 pKa = 10.6DD2493 pKa = 3.27STSAITAINSLVEE2506 pKa = 3.75GVYY2509 pKa = 9.98QFEE2512 pKa = 4.64LTVTDD2517 pKa = 3.41NDD2519 pKa = 3.82GAIAIDD2525 pKa = 3.8EE2526 pKa = 4.42VQVTVNAAPNIPPVANAGTDD2546 pKa = 3.65KK2547 pKa = 10.93IITLPTNLVTISGSGNDD2564 pKa = 3.46ADD2566 pKa = 4.19GTIVSYY2572 pKa = 10.74QWTKK2576 pKa = 10.53ISGPANYY2583 pKa = 9.97NILDD2587 pKa = 3.92ATTATIDD2594 pKa = 3.38INSLVEE2600 pKa = 4.06GVYY2603 pKa = 9.85QFQLKK2608 pKa = 8.09VTDD2611 pKa = 3.86NKK2613 pKa = 10.58GASATDD2619 pKa = 3.2IMQVTVNAAPNVPPVANAGPDD2640 pKa = 3.51QIITLPANTLTISGSGSDD2658 pKa = 3.46ADD2660 pKa = 4.0GTVVGYY2666 pKa = 10.46LWTKK2670 pKa = 10.11VSGPANYY2677 pKa = 10.28NVVNPTSSVTGINGLVQGVYY2697 pKa = 9.42QFQLKK2702 pKa = 8.35ITDD2705 pKa = 3.66NKK2707 pKa = 10.48GATSTDD2713 pKa = 2.56IMQVIVNAAANKK2725 pKa = 10.41APVANAGADD2734 pKa = 3.55KK2735 pKa = 10.98SITLPTNSLTVSGSGSDD2752 pKa = 3.16ADD2754 pKa = 4.79GIVASYY2760 pKa = 9.67QWSKK2764 pKa = 11.02ISGPTSYY2771 pKa = 11.13NIVNPTSAVTDD2782 pKa = 3.4INGLVQGVYY2791 pKa = 8.92QFEE2794 pKa = 4.84LRR2796 pKa = 11.84VTDD2799 pKa = 3.53NNGARR2804 pKa = 11.84GRR2806 pKa = 11.84DD2807 pKa = 3.42TVQITVNAAVNIPPIAHH2824 pKa = 7.57AGPDD2828 pKa = 3.27ITITLPVNTASLAGSGSDD2846 pKa = 3.26ADD2848 pKa = 4.04GTVVSYY2854 pKa = 11.1VWTKK2858 pKa = 10.18ISGPSTYY2865 pKa = 10.76NIINASSPVTDD2876 pKa = 2.59VWGLLKK2882 pKa = 10.49GVYY2885 pKa = 9.6KK2886 pKa = 10.28FEE2888 pKa = 6.71IIVTDD2893 pKa = 3.55NKK2895 pKa = 10.84GATSRR2900 pKa = 11.84DD2901 pKa = 3.28TMQITVNSATNIAPVANAGKK2921 pKa = 9.57DD2922 pKa = 3.4QTITLPTNSVSLTGSGTDD2940 pKa = 3.11ADD2942 pKa = 4.23GTIVGYY2948 pKa = 7.91SWKK2951 pKa = 10.39QISGPSASVIASTNSAKK2968 pKa = 10.02TSANGLVGGTYY2979 pKa = 9.46EE2980 pKa = 4.98FEE2982 pKa = 4.14LTVIDD2987 pKa = 3.83NLGAVGKK2994 pKa = 8.56DD2995 pKa = 3.05TVAIVVAEE3003 pKa = 3.93PRR3005 pKa = 11.84LNLNVQTNTLMACPNPVIDD3024 pKa = 4.2EE3025 pKa = 4.31TTLEE3029 pKa = 4.12IKK3031 pKa = 9.88TSQVYY3036 pKa = 10.54SKK3038 pKa = 11.06LLLIVTDD3045 pKa = 3.82MNGKK3049 pKa = 8.96IVYY3052 pKa = 9.57KK3053 pKa = 10.72KK3054 pKa = 10.14EE3055 pKa = 3.54ITGGQTEE3062 pKa = 4.7LKK3064 pKa = 10.22EE3065 pKa = 4.03KK3066 pKa = 11.12LNMSNLIKK3074 pKa = 9.88GTYY3077 pKa = 9.97AITIYY3082 pKa = 11.02YY3083 pKa = 9.36SDD3085 pKa = 4.96KK3086 pKa = 9.65EE3087 pKa = 4.13KK3088 pKa = 11.05QSIKK3092 pKa = 10.49VIRR3095 pKa = 11.84LL3096 pKa = 3.24
Molecular weight: 320.75 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5J5IM73|A0A5J5IM73_9BACT 7-carboxy-7-deazaguanine synthase OS=Ginsengibacter hankyongi OX=2607284 GN=queE PE=3 SV=1
MM1 pKa = 7.94RR2 pKa = 11.84EE3 pKa = 3.85RR4 pKa = 11.84IVRR7 pKa = 11.84AVAGAFVLTSIMLAYY22 pKa = 9.76FININWLWLGVFVGVNLLQSSFTRR46 pKa = 11.84FCPLEE51 pKa = 4.48NILNVLGVKK60 pKa = 8.45KK61 pKa = 10.07HH62 pKa = 5.24KK63 pKa = 9.88MKK65 pKa = 10.97NN66 pKa = 3.2
MM1 pKa = 7.94RR2 pKa = 11.84EE3 pKa = 3.85RR4 pKa = 11.84IVRR7 pKa = 11.84AVAGAFVLTSIMLAYY22 pKa = 9.76FININWLWLGVFVGVNLLQSSFTRR46 pKa = 11.84FCPLEE51 pKa = 4.48NILNVLGVKK60 pKa = 8.45KK61 pKa = 10.07HH62 pKa = 5.24KK63 pKa = 9.88MKK65 pKa = 10.97NN66 pKa = 3.2
Molecular weight: 7.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1583608 |
25 |
3096 |
353.8 |
39.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.814 ± 0.033 | 0.905 ± 0.011 |
5.302 ± 0.027 | 5.506 ± 0.04 |
5.132 ± 0.028 | 6.789 ± 0.034 |
1.909 ± 0.018 | 7.895 ± 0.033 |
7.567 ± 0.034 | 9.015 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.314 ± 0.019 | 6.205 ± 0.034 |
3.882 ± 0.02 | 3.421 ± 0.022 |
3.554 ± 0.021 | 6.55 ± 0.028 |
5.709 ± 0.035 | 6.081 ± 0.027 |
1.311 ± 0.015 | 4.139 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
