
Halanaeroarchaeum sulfurireducens
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Halobacteriales; Halobacteriaceae; Halanaeroarchaeum
Average proteome isoelectric point is 4.95
Get precalculated fractions of proteins
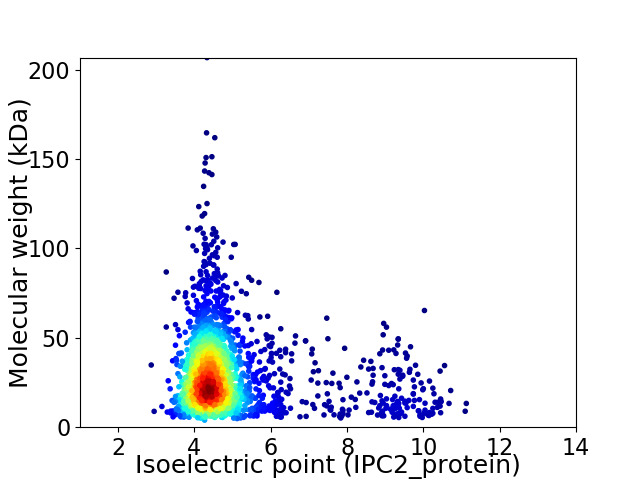
Virtual 2D-PAGE plot for 2225 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0F7PBN9|A0A0F7PBN9_9EURY Phosphoglycerate kinase OS=Halanaeroarchaeum sulfurireducens OX=1604004 GN=pgk PE=3 SV=1
MM1 pKa = 7.42GLSEE5 pKa = 4.17TLSTFFGFEE14 pKa = 3.74EE15 pKa = 5.07HH16 pKa = 7.15DD17 pKa = 3.31TDD19 pKa = 5.44LRR21 pKa = 11.84TEE23 pKa = 3.9ILAGVTTFLTMSYY36 pKa = 9.96IVVVNPAILTDD47 pKa = 3.54QPGDD51 pKa = 3.36GGFTDD56 pKa = 6.17GISLANHH63 pKa = 6.26SPGEE67 pKa = 4.19VQQMLAVVTILAASVAIFVMAFYY90 pKa = 11.02ANRR93 pKa = 11.84PFAQAPGLGLNAFFAFTVTGAMGIPWEE120 pKa = 4.13TALAAVFVEE129 pKa = 4.05GLVFILLTSVGARR142 pKa = 11.84EE143 pKa = 4.07YY144 pKa = 11.02VISLFPAPVKK154 pKa = 10.47FSVGTGIGLFLAIIGLQAMGIVAPDD179 pKa = 3.95EE180 pKa = 4.01ATMIAMGDD188 pKa = 3.84LAQDD192 pKa = 3.86PVAVLSVVGLLFTFALYY209 pKa = 10.79ARR211 pKa = 11.84GIPGAIVFGIIGTTIAGWALTAAGVTPPGVLAPEE245 pKa = 4.35SLPGAQYY252 pKa = 10.69DD253 pKa = 3.6ISPLAGAFIEE263 pKa = 5.16GFANVEE269 pKa = 4.01AFSFALVVFTFFFVDD284 pKa = 4.8FFDD287 pKa = 3.88TAGTLTGVSQVAGFLDD303 pKa = 4.75DD304 pKa = 4.96EE305 pKa = 5.23GNLPDD310 pKa = 4.68ADD312 pKa = 4.26KK313 pKa = 11.15PLMADD318 pKa = 4.37AIGTTVGSMLGTSTVTTFVEE338 pKa = 4.47SATGVEE344 pKa = 4.02AGGRR348 pKa = 11.84TGMTAFVVGVLFLLSLAIVPLAAAIPQYY376 pKa = 10.65ASHH379 pKa = 6.93IALVVVAVIMFANVTDD395 pKa = 4.1IQWDD399 pKa = 4.06DD400 pKa = 3.57LTHH403 pKa = 7.32AIPAGLTIIVMPFTYY418 pKa = 10.37SIAYY422 pKa = 8.97GIAAGIVSYY431 pKa = 9.62PIVKK435 pKa = 10.07SAVGEE440 pKa = 3.83YY441 pKa = 10.93DD442 pKa = 4.02EE443 pKa = 5.0IHH445 pKa = 5.83VGQWVLAAAFVLYY458 pKa = 9.41FFVRR462 pKa = 11.84TGGILTGAVV471 pKa = 3.18
MM1 pKa = 7.42GLSEE5 pKa = 4.17TLSTFFGFEE14 pKa = 3.74EE15 pKa = 5.07HH16 pKa = 7.15DD17 pKa = 3.31TDD19 pKa = 5.44LRR21 pKa = 11.84TEE23 pKa = 3.9ILAGVTTFLTMSYY36 pKa = 9.96IVVVNPAILTDD47 pKa = 3.54QPGDD51 pKa = 3.36GGFTDD56 pKa = 6.17GISLANHH63 pKa = 6.26SPGEE67 pKa = 4.19VQQMLAVVTILAASVAIFVMAFYY90 pKa = 11.02ANRR93 pKa = 11.84PFAQAPGLGLNAFFAFTVTGAMGIPWEE120 pKa = 4.13TALAAVFVEE129 pKa = 4.05GLVFILLTSVGARR142 pKa = 11.84EE143 pKa = 4.07YY144 pKa = 11.02VISLFPAPVKK154 pKa = 10.47FSVGTGIGLFLAIIGLQAMGIVAPDD179 pKa = 3.95EE180 pKa = 4.01ATMIAMGDD188 pKa = 3.84LAQDD192 pKa = 3.86PVAVLSVVGLLFTFALYY209 pKa = 10.79ARR211 pKa = 11.84GIPGAIVFGIIGTTIAGWALTAAGVTPPGVLAPEE245 pKa = 4.35SLPGAQYY252 pKa = 10.69DD253 pKa = 3.6ISPLAGAFIEE263 pKa = 5.16GFANVEE269 pKa = 4.01AFSFALVVFTFFFVDD284 pKa = 4.8FFDD287 pKa = 3.88TAGTLTGVSQVAGFLDD303 pKa = 4.75DD304 pKa = 4.96EE305 pKa = 5.23GNLPDD310 pKa = 4.68ADD312 pKa = 4.26KK313 pKa = 11.15PLMADD318 pKa = 4.37AIGTTVGSMLGTSTVTTFVEE338 pKa = 4.47SATGVEE344 pKa = 4.02AGGRR348 pKa = 11.84TGMTAFVVGVLFLLSLAIVPLAAAIPQYY376 pKa = 10.65ASHH379 pKa = 6.93IALVVVAVIMFANVTDD395 pKa = 4.1IQWDD399 pKa = 4.06DD400 pKa = 3.57LTHH403 pKa = 7.32AIPAGLTIIVMPFTYY418 pKa = 10.37SIAYY422 pKa = 8.97GIAAGIVSYY431 pKa = 9.62PIVKK435 pKa = 10.07SAVGEE440 pKa = 3.83YY441 pKa = 10.93DD442 pKa = 4.02EE443 pKa = 5.0IHH445 pKa = 5.83VGQWVLAAAFVLYY458 pKa = 9.41FFVRR462 pKa = 11.84TGGILTGAVV471 pKa = 3.18
Molecular weight: 48.78 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0F7P769|A0A0F7P769_9EURY UspA domain-containing protein OS=Halanaeroarchaeum sulfurireducens OX=1604004 GN=uspA1 PE=4 SV=1
MM1 pKa = 7.54SRR3 pKa = 11.84RR4 pKa = 11.84TPHH7 pKa = 6.1IPEE10 pKa = 3.97RR11 pKa = 11.84VRR13 pKa = 11.84RR14 pKa = 11.84GPTSSRR20 pKa = 11.84NSRR23 pKa = 11.84RR24 pKa = 11.84PARR27 pKa = 11.84FASVSSRR34 pKa = 11.84FRR36 pKa = 11.84PTTSTTPGPTKK47 pKa = 9.53RR48 pKa = 11.84HH49 pKa = 5.16SRR51 pKa = 11.84PTHH54 pKa = 5.42RR55 pKa = 11.84QNGSPVTNRR64 pKa = 11.84WPGAPASEE72 pKa = 4.46RR73 pKa = 11.84GPKK76 pKa = 8.12VTARR80 pKa = 11.84RR81 pKa = 11.84TAVSTPSRR89 pKa = 11.84RR90 pKa = 11.84FSRR93 pKa = 11.84TTRR96 pKa = 11.84RR97 pKa = 11.84SVRR100 pKa = 11.84FGTVLAPRR108 pKa = 11.84RR109 pKa = 11.84NGGPRR114 pKa = 11.84SGGSTTT120 pKa = 3.63
MM1 pKa = 7.54SRR3 pKa = 11.84RR4 pKa = 11.84TPHH7 pKa = 6.1IPEE10 pKa = 3.97RR11 pKa = 11.84VRR13 pKa = 11.84RR14 pKa = 11.84GPTSSRR20 pKa = 11.84NSRR23 pKa = 11.84RR24 pKa = 11.84PARR27 pKa = 11.84FASVSSRR34 pKa = 11.84FRR36 pKa = 11.84PTTSTTPGPTKK47 pKa = 9.53RR48 pKa = 11.84HH49 pKa = 5.16SRR51 pKa = 11.84PTHH54 pKa = 5.42RR55 pKa = 11.84QNGSPVTNRR64 pKa = 11.84WPGAPASEE72 pKa = 4.46RR73 pKa = 11.84GPKK76 pKa = 8.12VTARR80 pKa = 11.84RR81 pKa = 11.84TAVSTPSRR89 pKa = 11.84RR90 pKa = 11.84FSRR93 pKa = 11.84TTRR96 pKa = 11.84RR97 pKa = 11.84SVRR100 pKa = 11.84FGTVLAPRR108 pKa = 11.84RR109 pKa = 11.84NGGPRR114 pKa = 11.84SGGSTTT120 pKa = 3.63
Molecular weight: 13.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
637001 |
34 |
1872 |
286.3 |
31.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.45 ± 0.077 | 0.549 ± 0.017 |
8.407 ± 0.069 | 8.751 ± 0.079 |
3.242 ± 0.031 | 8.118 ± 0.049 |
2.058 ± 0.029 | 4.57 ± 0.042 |
2.066 ± 0.034 | 8.588 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.894 ± 0.02 | 2.416 ± 0.032 |
4.63 ± 0.038 | 2.593 ± 0.028 |
6.727 ± 0.055 | 5.534 ± 0.037 |
6.554 ± 0.044 | 9.15 ± 0.055 |
1.059 ± 0.022 | 2.643 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
