
Mesorhizobium sp. YR577
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Phyllobacteriaceae; Mesorhizobium; unclassified Mesorhizobium
Average proteome isoelectric point is 6.51
Get precalculated fractions of proteins
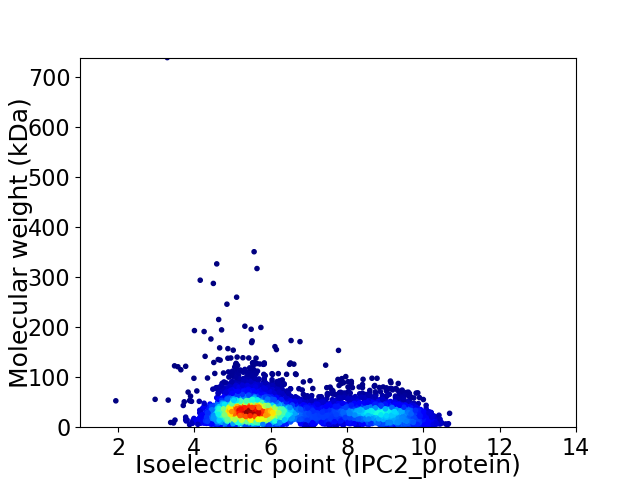
Virtual 2D-PAGE plot for 6444 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I7D715|A0A1I7D715_9RHIZ Crotonobetainyl-CoA hydratase OS=Mesorhizobium sp. YR577 OX=1884373 GN=SAMN05518861_111218 PE=3 SV=1
MM1 pKa = 6.86NTVLNAKK8 pKa = 8.71GVPLPYY14 pKa = 10.09SGSSVNHH21 pKa = 6.18FSATNSGPQLAGSALNDD38 pKa = 3.96SMWGDD43 pKa = 3.41SSVNVVMQGGTGDD56 pKa = 4.42DD57 pKa = 2.84IYY59 pKa = 11.62YY60 pKa = 10.35LYY62 pKa = 10.59SARR65 pKa = 11.84NSAFEE70 pKa = 3.96KK71 pKa = 10.56AGEE74 pKa = 4.29GVDD77 pKa = 5.09TINTWMSYY85 pKa = 10.37RR86 pKa = 11.84LPEE89 pKa = 4.5NIEE92 pKa = 3.93NLTVTGNGRR101 pKa = 11.84SAIGNDD107 pKa = 3.26GDD109 pKa = 4.73NILSGASGSQTIDD122 pKa = 2.68GGAGDD127 pKa = 4.95DD128 pKa = 3.76VLIGGGGADD137 pKa = 2.8IFVISEE143 pKa = 4.3GNGSDD148 pKa = 4.84LILDD152 pKa = 4.16FSVDD156 pKa = 3.47DD157 pKa = 3.73QVRR160 pKa = 11.84LDD162 pKa = 3.67GYY164 pKa = 11.11GFISFDD170 pKa = 4.09AIQSNMTQTGANVTLDD186 pKa = 3.79LGNDD190 pKa = 3.81EE191 pKa = 4.85ILVFANTTVDD201 pKa = 3.77QFDD204 pKa = 3.49AGQFKK209 pKa = 11.03LSLDD213 pKa = 3.54KK214 pKa = 11.58SEE216 pKa = 4.57MSLSFSDD223 pKa = 4.84DD224 pKa = 3.76FNSLSLWNGEE234 pKa = 4.05SGTWDD239 pKa = 3.43SNFWWGAEE247 pKa = 3.87NGSTHH252 pKa = 6.55EE253 pKa = 4.64GNGEE257 pKa = 4.01KK258 pKa = 9.65QWYY261 pKa = 9.12IDD263 pKa = 3.29TDD265 pKa = 3.88YY266 pKa = 11.83APTKK270 pKa = 10.1SVNPFSVDD278 pKa = 3.1NGVLTITAAPTPDD291 pKa = 4.15AIKK294 pKa = 10.72PEE296 pKa = 4.19VNNYY300 pKa = 10.76DD301 pKa = 3.71YY302 pKa = 11.4TSGLLTTYY310 pKa = 10.65EE311 pKa = 4.7SFSQTYY317 pKa = 10.09GYY319 pKa = 11.18FEE321 pKa = 4.44MRR323 pKa = 11.84ADD325 pKa = 3.44MPEE328 pKa = 3.97NQGVWPAFWLLPADD342 pKa = 4.39GSWPPEE348 pKa = 3.76LDD350 pKa = 3.52VVEE353 pKa = 4.68MRR355 pKa = 11.84GQEE358 pKa = 3.94PGVVHH363 pKa = 5.48VTSHH367 pKa = 5.1TNEE370 pKa = 4.46TGSHH374 pKa = 5.33TSVSSAVNVPDD385 pKa = 3.19TSGFHH390 pKa = 6.35TYY392 pKa = 9.8GVLWTEE398 pKa = 4.54EE399 pKa = 4.13EE400 pKa = 4.36IVWYY404 pKa = 10.3FDD406 pKa = 3.31DD407 pKa = 4.97VEE409 pKa = 4.25IARR412 pKa = 11.84TDD414 pKa = 3.66TPSDD418 pKa = 3.26MHH420 pKa = 8.21GPMYY424 pKa = 10.15MLVNLAVGGVAGTPADD440 pKa = 3.9GLATPAEE447 pKa = 4.02MQIDD451 pKa = 4.18YY452 pKa = 10.81IKK454 pKa = 10.54AYY456 pKa = 9.55EE457 pKa = 4.09LDD459 pKa = 3.71GVTQAAAKK467 pKa = 10.15AGSGDD472 pKa = 3.65FLVV475 pKa = 4.16
MM1 pKa = 6.86NTVLNAKK8 pKa = 8.71GVPLPYY14 pKa = 10.09SGSSVNHH21 pKa = 6.18FSATNSGPQLAGSALNDD38 pKa = 3.96SMWGDD43 pKa = 3.41SSVNVVMQGGTGDD56 pKa = 4.42DD57 pKa = 2.84IYY59 pKa = 11.62YY60 pKa = 10.35LYY62 pKa = 10.59SARR65 pKa = 11.84NSAFEE70 pKa = 3.96KK71 pKa = 10.56AGEE74 pKa = 4.29GVDD77 pKa = 5.09TINTWMSYY85 pKa = 10.37RR86 pKa = 11.84LPEE89 pKa = 4.5NIEE92 pKa = 3.93NLTVTGNGRR101 pKa = 11.84SAIGNDD107 pKa = 3.26GDD109 pKa = 4.73NILSGASGSQTIDD122 pKa = 2.68GGAGDD127 pKa = 4.95DD128 pKa = 3.76VLIGGGGADD137 pKa = 2.8IFVISEE143 pKa = 4.3GNGSDD148 pKa = 4.84LILDD152 pKa = 4.16FSVDD156 pKa = 3.47DD157 pKa = 3.73QVRR160 pKa = 11.84LDD162 pKa = 3.67GYY164 pKa = 11.11GFISFDD170 pKa = 4.09AIQSNMTQTGANVTLDD186 pKa = 3.79LGNDD190 pKa = 3.81EE191 pKa = 4.85ILVFANTTVDD201 pKa = 3.77QFDD204 pKa = 3.49AGQFKK209 pKa = 11.03LSLDD213 pKa = 3.54KK214 pKa = 11.58SEE216 pKa = 4.57MSLSFSDD223 pKa = 4.84DD224 pKa = 3.76FNSLSLWNGEE234 pKa = 4.05SGTWDD239 pKa = 3.43SNFWWGAEE247 pKa = 3.87NGSTHH252 pKa = 6.55EE253 pKa = 4.64GNGEE257 pKa = 4.01KK258 pKa = 9.65QWYY261 pKa = 9.12IDD263 pKa = 3.29TDD265 pKa = 3.88YY266 pKa = 11.83APTKK270 pKa = 10.1SVNPFSVDD278 pKa = 3.1NGVLTITAAPTPDD291 pKa = 4.15AIKK294 pKa = 10.72PEE296 pKa = 4.19VNNYY300 pKa = 10.76DD301 pKa = 3.71YY302 pKa = 11.4TSGLLTTYY310 pKa = 10.65EE311 pKa = 4.7SFSQTYY317 pKa = 10.09GYY319 pKa = 11.18FEE321 pKa = 4.44MRR323 pKa = 11.84ADD325 pKa = 3.44MPEE328 pKa = 3.97NQGVWPAFWLLPADD342 pKa = 4.39GSWPPEE348 pKa = 3.76LDD350 pKa = 3.52VVEE353 pKa = 4.68MRR355 pKa = 11.84GQEE358 pKa = 3.94PGVVHH363 pKa = 5.48VTSHH367 pKa = 5.1TNEE370 pKa = 4.46TGSHH374 pKa = 5.33TSVSSAVNVPDD385 pKa = 3.19TSGFHH390 pKa = 6.35TYY392 pKa = 9.8GVLWTEE398 pKa = 4.54EE399 pKa = 4.13EE400 pKa = 4.36IVWYY404 pKa = 10.3FDD406 pKa = 3.31DD407 pKa = 4.97VEE409 pKa = 4.25IARR412 pKa = 11.84TDD414 pKa = 3.66TPSDD418 pKa = 3.26MHH420 pKa = 8.21GPMYY424 pKa = 10.15MLVNLAVGGVAGTPADD440 pKa = 3.9GLATPAEE447 pKa = 4.02MQIDD451 pKa = 4.18YY452 pKa = 10.81IKK454 pKa = 10.54AYY456 pKa = 9.55EE457 pKa = 4.09LDD459 pKa = 3.71GVTQAAAKK467 pKa = 10.15AGSGDD472 pKa = 3.65FLVV475 pKa = 4.16
Molecular weight: 50.81 kDa
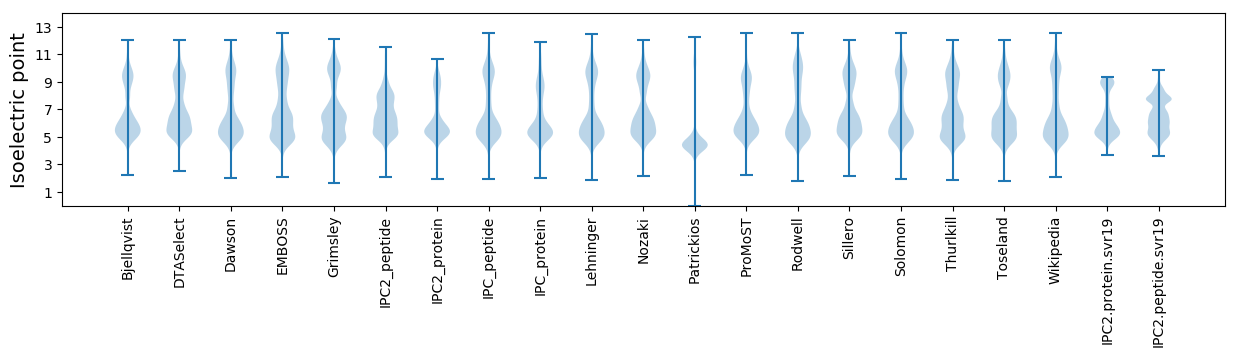
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I6ZVZ7|A0A1I6ZVZ7_9RHIZ Manganese/iron transport system permease protein OS=Mesorhizobium sp. YR577 OX=1884373 GN=SAMN05518861_103255 PE=3 SV=1
MM1 pKa = 8.09DD2 pKa = 4.95KK3 pKa = 11.06GDD5 pKa = 3.98RR6 pKa = 11.84RR7 pKa = 11.84PLASRR12 pKa = 11.84NTRR15 pKa = 11.84WAQTIAKK22 pKa = 9.45RR23 pKa = 11.84MAALSITPNAISQASIAAAAVAGLAFWIAGEE54 pKa = 4.17TSGTARR60 pKa = 11.84SLLLVAAALFCQLRR74 pKa = 11.84LLCNLFDD81 pKa = 3.98GMVAVEE87 pKa = 4.26GGKK90 pKa = 9.78AAADD94 pKa = 3.84GPFWNEE100 pKa = 3.4FPDD103 pKa = 4.88RR104 pKa = 11.84IADD107 pKa = 3.62ILILAGVGYY116 pKa = 10.69GIGAPSLGWAAAAFAVLTAYY136 pKa = 10.13VRR138 pKa = 11.84EE139 pKa = 4.14LGRR142 pKa = 11.84ANGAPSDD149 pKa = 4.12FSGPMAKK156 pKa = 7.72QHH158 pKa = 5.49RR159 pKa = 11.84MAVITAAALFSAVEE173 pKa = 4.12FLWHH177 pKa = 6.23GRR179 pKa = 11.84NEE181 pKa = 4.25VLTLALSIVAIGASLTAFRR200 pKa = 11.84RR201 pKa = 11.84ARR203 pKa = 11.84RR204 pKa = 11.84QVRR207 pKa = 11.84WLKK210 pKa = 9.88EE211 pKa = 3.66RR212 pKa = 11.84KK213 pKa = 8.84AWQSTPP219 pKa = 3.1
MM1 pKa = 8.09DD2 pKa = 4.95KK3 pKa = 11.06GDD5 pKa = 3.98RR6 pKa = 11.84RR7 pKa = 11.84PLASRR12 pKa = 11.84NTRR15 pKa = 11.84WAQTIAKK22 pKa = 9.45RR23 pKa = 11.84MAALSITPNAISQASIAAAAVAGLAFWIAGEE54 pKa = 4.17TSGTARR60 pKa = 11.84SLLLVAAALFCQLRR74 pKa = 11.84LLCNLFDD81 pKa = 3.98GMVAVEE87 pKa = 4.26GGKK90 pKa = 9.78AAADD94 pKa = 3.84GPFWNEE100 pKa = 3.4FPDD103 pKa = 4.88RR104 pKa = 11.84IADD107 pKa = 3.62ILILAGVGYY116 pKa = 10.69GIGAPSLGWAAAAFAVLTAYY136 pKa = 10.13VRR138 pKa = 11.84EE139 pKa = 4.14LGRR142 pKa = 11.84ANGAPSDD149 pKa = 4.12FSGPMAKK156 pKa = 7.72QHH158 pKa = 5.49RR159 pKa = 11.84MAVITAAALFSAVEE173 pKa = 4.12FLWHH177 pKa = 6.23GRR179 pKa = 11.84NEE181 pKa = 4.25VLTLALSIVAIGASLTAFRR200 pKa = 11.84RR201 pKa = 11.84ARR203 pKa = 11.84RR204 pKa = 11.84QVRR207 pKa = 11.84WLKK210 pKa = 9.88EE211 pKa = 3.66RR212 pKa = 11.84KK213 pKa = 8.84AWQSTPP219 pKa = 3.1
Molecular weight: 23.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2014053 |
26 |
7420 |
312.5 |
33.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.005 ± 0.04 | 0.786 ± 0.009 |
5.633 ± 0.031 | 5.705 ± 0.032 |
3.951 ± 0.022 | 8.592 ± 0.039 |
1.953 ± 0.016 | 5.633 ± 0.027 |
3.742 ± 0.026 | 9.868 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.56 ± 0.017 | 2.835 ± 0.025 |
4.976 ± 0.022 | 2.988 ± 0.016 |
6.561 ± 0.036 | 5.725 ± 0.025 |
5.421 ± 0.046 | 7.466 ± 0.025 |
1.314 ± 0.012 | 2.286 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
