
Hubei sobemo-like virus 20
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.84
Get precalculated fractions of proteins
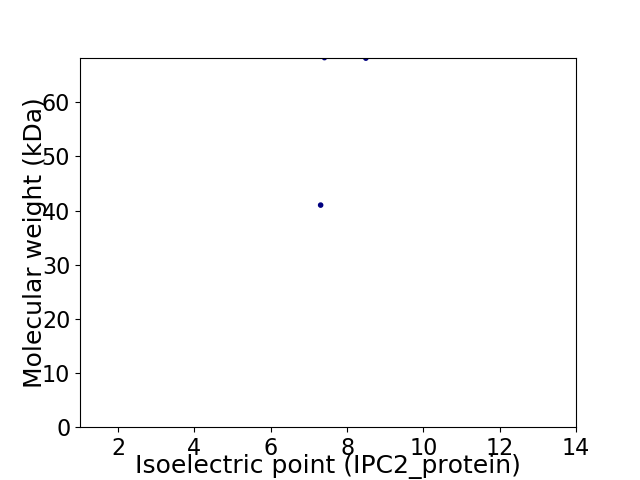
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
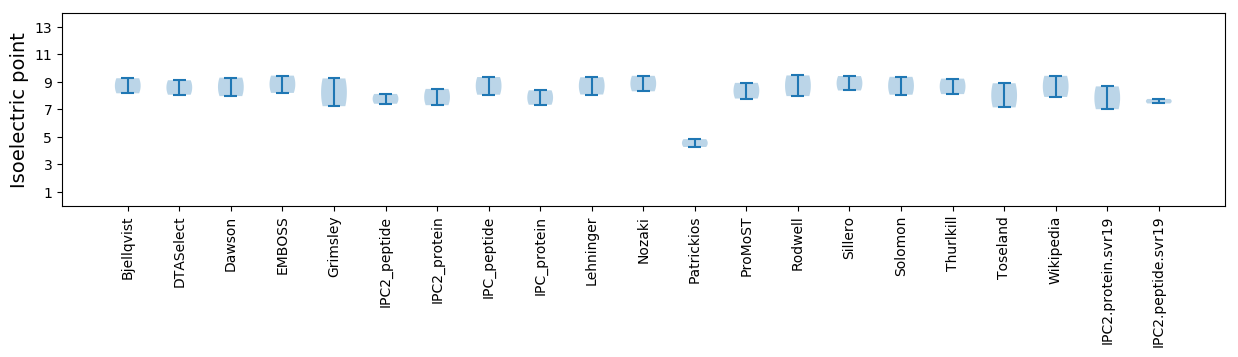
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEC2|A0A1L3KEC2_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 20 OX=1923206 PE=4 SV=1
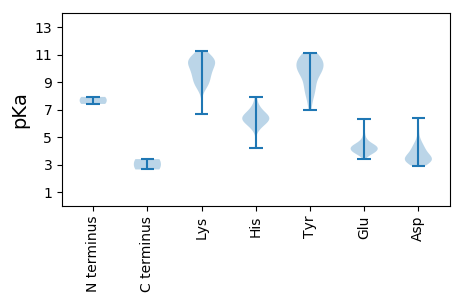
MM1 pKa = 7.92RR2 pKa = 11.84ALQRR6 pKa = 11.84LDD8 pKa = 4.62LNSSPGYY15 pKa = 9.61PYY17 pKa = 10.83LRR19 pKa = 11.84QATNNRR25 pKa = 11.84QLFTDD30 pKa = 3.6VDD32 pKa = 4.72GNWLPDD38 pKa = 3.37RR39 pKa = 11.84VEE41 pKa = 3.98WCWDD45 pKa = 3.3LVSRR49 pKa = 11.84RR50 pKa = 11.84LGNLSSADD58 pKa = 4.02PIRR61 pKa = 11.84LFVKK65 pKa = 10.16PEE67 pKa = 3.6PHH69 pKa = 6.4SLKK72 pKa = 10.58KK73 pKa = 9.76MHH75 pKa = 7.25DD76 pKa = 2.93KK77 pKa = 10.48RR78 pKa = 11.84YY79 pKa = 9.87RR80 pKa = 11.84LISSVSVVDD89 pKa = 4.35QIIDD93 pKa = 3.32HH94 pKa = 6.57MIFGDD99 pKa = 5.24LNDD102 pKa = 4.55LLIKK106 pKa = 9.57NWHH109 pKa = 6.35VIPSKK114 pKa = 10.35PGWSFLHH121 pKa = 6.56GGWKK125 pKa = 9.13MMPPKK130 pKa = 10.11GKK132 pKa = 8.94WVAIDD137 pKa = 3.46KK138 pKa = 10.87SSWDD142 pKa = 3.16WTVQLWLLEE151 pKa = 4.2MVLEE155 pKa = 4.37ARR157 pKa = 11.84LRR159 pKa = 11.84LCRR162 pKa = 11.84TTGEE166 pKa = 4.19LRR168 pKa = 11.84NRR170 pKa = 11.84WFKK173 pKa = 8.83YY174 pKa = 8.94AYY176 pKa = 10.14YY177 pKa = 10.34RR178 pKa = 11.84FEE180 pKa = 4.05QLYY183 pKa = 8.81LKK185 pKa = 10.62CVFITSGGHH194 pKa = 6.58LLRR197 pKa = 11.84QLRR200 pKa = 11.84PGALKK205 pKa = 10.42SGCVNTIADD214 pKa = 4.17NSLMQYY220 pKa = 10.53LLHH223 pKa = 6.41LRR225 pKa = 11.84VVFQLGYY232 pKa = 9.76SPGIIMSMGDD242 pKa = 3.25DD243 pKa = 3.61TLQQEE248 pKa = 4.59PEE250 pKa = 3.92DD251 pKa = 3.66LQRR254 pKa = 11.84YY255 pKa = 6.96IQEE258 pKa = 4.26LQTFCIVKK266 pKa = 9.39QYY268 pKa = 10.49EE269 pKa = 4.2RR270 pKa = 11.84LVEE273 pKa = 3.92FAGFRR278 pKa = 11.84FDD280 pKa = 4.19GMQVEE285 pKa = 4.35PSYY288 pKa = 10.38RR289 pKa = 11.84AKK291 pKa = 10.7HH292 pKa = 5.61SFNLLHH298 pKa = 7.14VDD300 pKa = 3.91DD301 pKa = 5.74AVLPDD306 pKa = 3.37VAVAYY311 pKa = 10.84SLMYY315 pKa = 10.0HH316 pKa = 6.21RR317 pKa = 11.84SKK319 pKa = 10.43YY320 pKa = 9.77RR321 pKa = 11.84VWIRR325 pKa = 11.84SLFEE329 pKa = 4.1EE330 pKa = 4.68MGLDD334 pKa = 4.04LPSLEE339 pKa = 4.19YY340 pKa = 10.47FDD342 pKa = 6.4AIFDD346 pKa = 4.2GEE348 pKa = 4.43VV349 pKa = 2.64
MM1 pKa = 7.92RR2 pKa = 11.84ALQRR6 pKa = 11.84LDD8 pKa = 4.62LNSSPGYY15 pKa = 9.61PYY17 pKa = 10.83LRR19 pKa = 11.84QATNNRR25 pKa = 11.84QLFTDD30 pKa = 3.6VDD32 pKa = 4.72GNWLPDD38 pKa = 3.37RR39 pKa = 11.84VEE41 pKa = 3.98WCWDD45 pKa = 3.3LVSRR49 pKa = 11.84RR50 pKa = 11.84LGNLSSADD58 pKa = 4.02PIRR61 pKa = 11.84LFVKK65 pKa = 10.16PEE67 pKa = 3.6PHH69 pKa = 6.4SLKK72 pKa = 10.58KK73 pKa = 9.76MHH75 pKa = 7.25DD76 pKa = 2.93KK77 pKa = 10.48RR78 pKa = 11.84YY79 pKa = 9.87RR80 pKa = 11.84LISSVSVVDD89 pKa = 4.35QIIDD93 pKa = 3.32HH94 pKa = 6.57MIFGDD99 pKa = 5.24LNDD102 pKa = 4.55LLIKK106 pKa = 9.57NWHH109 pKa = 6.35VIPSKK114 pKa = 10.35PGWSFLHH121 pKa = 6.56GGWKK125 pKa = 9.13MMPPKK130 pKa = 10.11GKK132 pKa = 8.94WVAIDD137 pKa = 3.46KK138 pKa = 10.87SSWDD142 pKa = 3.16WTVQLWLLEE151 pKa = 4.2MVLEE155 pKa = 4.37ARR157 pKa = 11.84LRR159 pKa = 11.84LCRR162 pKa = 11.84TTGEE166 pKa = 4.19LRR168 pKa = 11.84NRR170 pKa = 11.84WFKK173 pKa = 8.83YY174 pKa = 8.94AYY176 pKa = 10.14YY177 pKa = 10.34RR178 pKa = 11.84FEE180 pKa = 4.05QLYY183 pKa = 8.81LKK185 pKa = 10.62CVFITSGGHH194 pKa = 6.58LLRR197 pKa = 11.84QLRR200 pKa = 11.84PGALKK205 pKa = 10.42SGCVNTIADD214 pKa = 4.17NSLMQYY220 pKa = 10.53LLHH223 pKa = 6.41LRR225 pKa = 11.84VVFQLGYY232 pKa = 9.76SPGIIMSMGDD242 pKa = 3.25DD243 pKa = 3.61TLQQEE248 pKa = 4.59PEE250 pKa = 3.92DD251 pKa = 3.66LQRR254 pKa = 11.84YY255 pKa = 6.96IQEE258 pKa = 4.26LQTFCIVKK266 pKa = 9.39QYY268 pKa = 10.49EE269 pKa = 4.2RR270 pKa = 11.84LVEE273 pKa = 3.92FAGFRR278 pKa = 11.84FDD280 pKa = 4.19GMQVEE285 pKa = 4.35PSYY288 pKa = 10.38RR289 pKa = 11.84AKK291 pKa = 10.7HH292 pKa = 5.61SFNLLHH298 pKa = 7.14VDD300 pKa = 3.91DD301 pKa = 5.74AVLPDD306 pKa = 3.37VAVAYY311 pKa = 10.84SLMYY315 pKa = 10.0HH316 pKa = 6.21RR317 pKa = 11.84SKK319 pKa = 10.43YY320 pKa = 9.77RR321 pKa = 11.84VWIRR325 pKa = 11.84SLFEE329 pKa = 4.1EE330 pKa = 4.68MGLDD334 pKa = 4.04LPSLEE339 pKa = 4.19YY340 pKa = 10.47FDD342 pKa = 6.4AIFDD346 pKa = 4.2GEE348 pKa = 4.43VV349 pKa = 2.64
Molecular weight: 41.01 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEC2|A0A1L3KEC2_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 20 OX=1923206 PE=4 SV=1
MM1 pKa = 7.41EE2 pKa = 6.3AEE4 pKa = 4.0QQLQQEE10 pKa = 4.85KK11 pKa = 10.25YY12 pKa = 9.65LLDD15 pKa = 3.64VMVDD19 pKa = 3.04KK20 pKa = 10.71VTVIGRR26 pKa = 11.84RR27 pKa = 11.84IMMAYY32 pKa = 9.45KK33 pKa = 9.39LTVNAVFLFSVYY45 pKa = 9.69SWICRR50 pKa = 11.84HH51 pKa = 5.96KK52 pKa = 11.11EE53 pKa = 3.42MVIRR57 pKa = 11.84PKK59 pKa = 10.57SRR61 pKa = 11.84YY62 pKa = 7.68EE63 pKa = 3.9LLWEE67 pKa = 3.93LVIEE71 pKa = 4.64HH72 pKa = 6.11SQPLVVVGVVVASIVVALLVRR93 pKa = 11.84RR94 pKa = 11.84SRR96 pKa = 11.84RR97 pKa = 11.84PVRR100 pKa = 11.84RR101 pKa = 11.84AIMRR105 pKa = 11.84LRR107 pKa = 11.84GVSLEE112 pKa = 3.8AMRR115 pKa = 11.84EE116 pKa = 4.03GSDD119 pKa = 4.21FRR121 pKa = 11.84AGDD124 pKa = 3.39QPKK127 pKa = 9.63FQVAILRR134 pKa = 11.84PGLLSDD140 pKa = 3.03SHH142 pKa = 7.92YY143 pKa = 10.98GYY145 pKa = 10.8GIRR148 pKa = 11.84VGDD151 pKa = 3.64FLVTPEE157 pKa = 4.34HH158 pKa = 6.33VVAGMPEE165 pKa = 4.33VILSANGNKK174 pKa = 9.03VQVPTVPMLRR184 pKa = 11.84SRR186 pKa = 11.84AFSDD190 pKa = 3.37LVYY193 pKa = 10.71IRR195 pKa = 11.84VSDD198 pKa = 4.08VAWSRR203 pKa = 11.84LAIRR207 pKa = 11.84TAVLSQKK214 pKa = 9.16TMATNLFVSVYY225 pKa = 7.93GQKK228 pKa = 10.56GVSSGLLSKK237 pKa = 10.85SQMVGLLVYY246 pKa = 10.36HH247 pKa = 7.03GSTVPGMSGAAYY259 pKa = 8.19TVGNAVYY266 pKa = 9.8GIHH269 pKa = 6.7CGVVANEE276 pKa = 3.92NMGVASLLLAAEE288 pKa = 5.12LKK290 pKa = 10.4RR291 pKa = 11.84YY292 pKa = 8.48MIGEE296 pKa = 4.07ASEE299 pKa = 5.38DD300 pKa = 3.71FAAQDD305 pKa = 2.96IWKK308 pKa = 9.03SQPKK312 pKa = 8.35KK313 pKa = 8.65TWDD316 pKa = 3.29SSDD319 pKa = 3.15VFKK322 pKa = 11.28VVEE325 pKa = 4.16RR326 pKa = 11.84RR327 pKa = 11.84FKK329 pKa = 10.62YY330 pKa = 8.82TDD332 pKa = 3.32EE333 pKa = 4.43EE334 pKa = 4.11QGEE337 pKa = 4.32MLARR341 pKa = 11.84GATWGQVCEE350 pKa = 4.18WADD353 pKa = 3.55EE354 pKa = 4.59SIAGAMATKK363 pKa = 10.01PKK365 pKa = 9.17TVKK368 pKa = 9.95PHH370 pKa = 4.23VTIEE374 pKa = 4.31DD375 pKa = 3.37GTIAMKK381 pKa = 8.62MHH383 pKa = 6.45PQGGSEE389 pKa = 4.28SGEE392 pKa = 3.91EE393 pKa = 3.86MTILAQLPAGASPKK407 pKa = 10.61ASNTYY412 pKa = 8.47QAEE415 pKa = 4.08EE416 pKa = 4.39VIADD420 pKa = 3.92LVRR423 pKa = 11.84RR424 pKa = 11.84VLEE427 pKa = 3.81LEE429 pKa = 3.69RR430 pKa = 11.84RR431 pKa = 11.84MDD433 pKa = 3.74RR434 pKa = 11.84VEE436 pKa = 4.14HH437 pKa = 6.23PMVEE441 pKa = 4.35VPLPEE446 pKa = 4.87AKK448 pKa = 10.07RR449 pKa = 11.84QFYY452 pKa = 10.28SCTMCSGKK460 pKa = 8.73FTDD463 pKa = 4.57PVAHH467 pKa = 6.75ARR469 pKa = 11.84LHH471 pKa = 6.02EE472 pKa = 4.43KK473 pKa = 9.88PICPQCGISCTDD485 pKa = 3.17EE486 pKa = 3.87QAVRR490 pKa = 11.84NHH492 pKa = 5.68LRR494 pKa = 11.84ASHH497 pKa = 5.93PQRR500 pKa = 11.84IQCDD504 pKa = 3.46RR505 pKa = 11.84CGVTCRR511 pKa = 11.84SSEE514 pKa = 3.69RR515 pKa = 11.84LARR518 pKa = 11.84HH519 pKa = 6.3RR520 pKa = 11.84KK521 pKa = 6.66TCIPMTDD528 pKa = 3.01GSFTKK533 pKa = 10.48EE534 pKa = 3.77SADD537 pKa = 3.38QVDD540 pKa = 3.31SRR542 pKa = 11.84KK543 pKa = 9.67VVRR546 pKa = 11.84TAGAFLGKK554 pKa = 9.71KK555 pKa = 9.73ASRR558 pKa = 11.84ARR560 pKa = 11.84RR561 pKa = 11.84SASSTRR567 pKa = 11.84SSSSSDD573 pKa = 3.01KK574 pKa = 11.26SPPCPSRR581 pKa = 11.84QDD583 pKa = 3.56YY584 pKa = 11.1LKK586 pKa = 10.92LILEE590 pKa = 4.63SQRR593 pKa = 11.84STQLALEE600 pKa = 4.41NLLSSMTGPSSGILQSS616 pKa = 3.4
MM1 pKa = 7.41EE2 pKa = 6.3AEE4 pKa = 4.0QQLQQEE10 pKa = 4.85KK11 pKa = 10.25YY12 pKa = 9.65LLDD15 pKa = 3.64VMVDD19 pKa = 3.04KK20 pKa = 10.71VTVIGRR26 pKa = 11.84RR27 pKa = 11.84IMMAYY32 pKa = 9.45KK33 pKa = 9.39LTVNAVFLFSVYY45 pKa = 9.69SWICRR50 pKa = 11.84HH51 pKa = 5.96KK52 pKa = 11.11EE53 pKa = 3.42MVIRR57 pKa = 11.84PKK59 pKa = 10.57SRR61 pKa = 11.84YY62 pKa = 7.68EE63 pKa = 3.9LLWEE67 pKa = 3.93LVIEE71 pKa = 4.64HH72 pKa = 6.11SQPLVVVGVVVASIVVALLVRR93 pKa = 11.84RR94 pKa = 11.84SRR96 pKa = 11.84RR97 pKa = 11.84PVRR100 pKa = 11.84RR101 pKa = 11.84AIMRR105 pKa = 11.84LRR107 pKa = 11.84GVSLEE112 pKa = 3.8AMRR115 pKa = 11.84EE116 pKa = 4.03GSDD119 pKa = 4.21FRR121 pKa = 11.84AGDD124 pKa = 3.39QPKK127 pKa = 9.63FQVAILRR134 pKa = 11.84PGLLSDD140 pKa = 3.03SHH142 pKa = 7.92YY143 pKa = 10.98GYY145 pKa = 10.8GIRR148 pKa = 11.84VGDD151 pKa = 3.64FLVTPEE157 pKa = 4.34HH158 pKa = 6.33VVAGMPEE165 pKa = 4.33VILSANGNKK174 pKa = 9.03VQVPTVPMLRR184 pKa = 11.84SRR186 pKa = 11.84AFSDD190 pKa = 3.37LVYY193 pKa = 10.71IRR195 pKa = 11.84VSDD198 pKa = 4.08VAWSRR203 pKa = 11.84LAIRR207 pKa = 11.84TAVLSQKK214 pKa = 9.16TMATNLFVSVYY225 pKa = 7.93GQKK228 pKa = 10.56GVSSGLLSKK237 pKa = 10.85SQMVGLLVYY246 pKa = 10.36HH247 pKa = 7.03GSTVPGMSGAAYY259 pKa = 8.19TVGNAVYY266 pKa = 9.8GIHH269 pKa = 6.7CGVVANEE276 pKa = 3.92NMGVASLLLAAEE288 pKa = 5.12LKK290 pKa = 10.4RR291 pKa = 11.84YY292 pKa = 8.48MIGEE296 pKa = 4.07ASEE299 pKa = 5.38DD300 pKa = 3.71FAAQDD305 pKa = 2.96IWKK308 pKa = 9.03SQPKK312 pKa = 8.35KK313 pKa = 8.65TWDD316 pKa = 3.29SSDD319 pKa = 3.15VFKK322 pKa = 11.28VVEE325 pKa = 4.16RR326 pKa = 11.84RR327 pKa = 11.84FKK329 pKa = 10.62YY330 pKa = 8.82TDD332 pKa = 3.32EE333 pKa = 4.43EE334 pKa = 4.11QGEE337 pKa = 4.32MLARR341 pKa = 11.84GATWGQVCEE350 pKa = 4.18WADD353 pKa = 3.55EE354 pKa = 4.59SIAGAMATKK363 pKa = 10.01PKK365 pKa = 9.17TVKK368 pKa = 9.95PHH370 pKa = 4.23VTIEE374 pKa = 4.31DD375 pKa = 3.37GTIAMKK381 pKa = 8.62MHH383 pKa = 6.45PQGGSEE389 pKa = 4.28SGEE392 pKa = 3.91EE393 pKa = 3.86MTILAQLPAGASPKK407 pKa = 10.61ASNTYY412 pKa = 8.47QAEE415 pKa = 4.08EE416 pKa = 4.39VIADD420 pKa = 3.92LVRR423 pKa = 11.84RR424 pKa = 11.84VLEE427 pKa = 3.81LEE429 pKa = 3.69RR430 pKa = 11.84RR431 pKa = 11.84MDD433 pKa = 3.74RR434 pKa = 11.84VEE436 pKa = 4.14HH437 pKa = 6.23PMVEE441 pKa = 4.35VPLPEE446 pKa = 4.87AKK448 pKa = 10.07RR449 pKa = 11.84QFYY452 pKa = 10.28SCTMCSGKK460 pKa = 8.73FTDD463 pKa = 4.57PVAHH467 pKa = 6.75ARR469 pKa = 11.84LHH471 pKa = 6.02EE472 pKa = 4.43KK473 pKa = 9.88PICPQCGISCTDD485 pKa = 3.17EE486 pKa = 3.87QAVRR490 pKa = 11.84NHH492 pKa = 5.68LRR494 pKa = 11.84ASHH497 pKa = 5.93PQRR500 pKa = 11.84IQCDD504 pKa = 3.46RR505 pKa = 11.84CGVTCRR511 pKa = 11.84SSEE514 pKa = 3.69RR515 pKa = 11.84LARR518 pKa = 11.84HH519 pKa = 6.3RR520 pKa = 11.84KK521 pKa = 6.66TCIPMTDD528 pKa = 3.01GSFTKK533 pKa = 10.48EE534 pKa = 3.77SADD537 pKa = 3.38QVDD540 pKa = 3.31SRR542 pKa = 11.84KK543 pKa = 9.67VVRR546 pKa = 11.84TAGAFLGKK554 pKa = 9.71KK555 pKa = 9.73ASRR558 pKa = 11.84ARR560 pKa = 11.84RR561 pKa = 11.84SASSTRR567 pKa = 11.84SSSSSDD573 pKa = 3.01KK574 pKa = 11.26SPPCPSRR581 pKa = 11.84QDD583 pKa = 3.56YY584 pKa = 11.1LKK586 pKa = 10.92LILEE590 pKa = 4.63SQRR593 pKa = 11.84STQLALEE600 pKa = 4.41NLLSSMTGPSSGILQSS616 pKa = 3.4
Molecular weight: 68.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
965 |
349 |
616 |
482.5 |
54.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.839 ± 1.608 | 1.865 ± 0.246 |
5.078 ± 1.023 | 5.596 ± 0.575 |
3.109 ± 0.839 | 6.218 ± 0.277 |
2.487 ± 0.215 | 4.352 ± 0.132 |
4.87 ± 0.163 | 9.948 ± 1.838 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.731 ± 0.166 | 2.176 ± 0.555 |
4.56 ± 0.014 | 4.56 ± 0.014 |
7.772 ± 0.183 | 8.705 ± 1.039 |
3.938 ± 0.773 | 8.912 ± 1.157 |
1.969 ± 0.835 | 3.316 ± 0.721 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
