
Pontibacter lucknowensis
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Hymenobacteraceae; Pontibacter
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
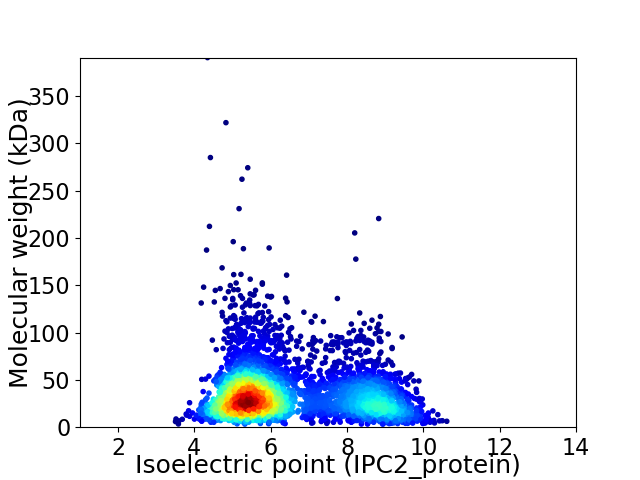
Virtual 2D-PAGE plot for 3901 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1N6U5R2|A0A1N6U5R2_9BACT ATP-binding cassette subfamily B OS=Pontibacter lucknowensis OX=1077936 GN=SAMN05421545_0696 PE=4 SV=1
MM1 pKa = 8.3DD2 pKa = 4.54SPQYY6 pKa = 8.75TPYY9 pKa = 10.5YY10 pKa = 8.81IQINEE15 pKa = 4.36LYY17 pKa = 9.55IDD19 pKa = 3.83TVVIDD24 pKa = 3.78DD25 pKa = 4.47TEE27 pKa = 5.88FITLWCFTVLDD38 pKa = 4.34DD39 pKa = 4.5EE40 pKa = 5.08YY41 pKa = 11.64KK42 pKa = 10.59FIYY45 pKa = 9.67LGCDD49 pKa = 3.38FTQFNQILIGAGEE62 pKa = 4.16KK63 pKa = 10.48GKK65 pKa = 10.71EE66 pKa = 4.1IALHH70 pKa = 5.24MANVLNSEE78 pKa = 4.3HH79 pKa = 7.55DD80 pKa = 3.87SPTLVPIKK88 pKa = 10.32EE89 pKa = 4.06QFDD92 pKa = 4.28DD93 pKa = 4.06YY94 pKa = 11.96LEE96 pKa = 4.21LTGFDD101 pKa = 3.52LVVYY105 pKa = 9.81EE106 pKa = 4.64PLAVDD111 pKa = 5.6DD112 pKa = 4.63EE113 pKa = 5.19FEE115 pKa = 5.55DD116 pKa = 5.62EE117 pKa = 5.34YY118 pKa = 11.64DD119 pKa = 5.39DD120 pKa = 6.78DD121 pKa = 6.78DD122 pKa = 5.86DD123 pKa = 6.48LPLEE127 pKa = 3.97QLLGGGSKK135 pKa = 10.57RR136 pKa = 11.84SDD138 pKa = 3.24NTSQKK143 pKa = 10.49YY144 pKa = 10.43LIYY147 pKa = 10.44QLDD150 pKa = 3.44KK151 pKa = 10.87LYY153 pKa = 9.91PSKK156 pKa = 11.07VFEE159 pKa = 4.22PEE161 pKa = 3.34KK162 pKa = 10.3LQRR165 pKa = 11.84EE166 pKa = 4.25NMTEE170 pKa = 3.88QFEE173 pKa = 4.56DD174 pKa = 4.14DD175 pKa = 4.12SLEE178 pKa = 4.17LAQLYY183 pKa = 10.35YY184 pKa = 10.83DD185 pKa = 3.96YY186 pKa = 11.63LNASEE191 pKa = 5.01NGYY194 pKa = 8.58SEE196 pKa = 5.04CEE198 pKa = 3.74ARR200 pKa = 11.84QVAGLEE206 pKa = 3.73KK207 pKa = 10.92DD208 pKa = 3.52LFFRR212 pKa = 11.84MARR215 pKa = 11.84KK216 pKa = 8.92AADD219 pKa = 3.13LWSS222 pKa = 3.58
MM1 pKa = 8.3DD2 pKa = 4.54SPQYY6 pKa = 8.75TPYY9 pKa = 10.5YY10 pKa = 8.81IQINEE15 pKa = 4.36LYY17 pKa = 9.55IDD19 pKa = 3.83TVVIDD24 pKa = 3.78DD25 pKa = 4.47TEE27 pKa = 5.88FITLWCFTVLDD38 pKa = 4.34DD39 pKa = 4.5EE40 pKa = 5.08YY41 pKa = 11.64KK42 pKa = 10.59FIYY45 pKa = 9.67LGCDD49 pKa = 3.38FTQFNQILIGAGEE62 pKa = 4.16KK63 pKa = 10.48GKK65 pKa = 10.71EE66 pKa = 4.1IALHH70 pKa = 5.24MANVLNSEE78 pKa = 4.3HH79 pKa = 7.55DD80 pKa = 3.87SPTLVPIKK88 pKa = 10.32EE89 pKa = 4.06QFDD92 pKa = 4.28DD93 pKa = 4.06YY94 pKa = 11.96LEE96 pKa = 4.21LTGFDD101 pKa = 3.52LVVYY105 pKa = 9.81EE106 pKa = 4.64PLAVDD111 pKa = 5.6DD112 pKa = 4.63EE113 pKa = 5.19FEE115 pKa = 5.55DD116 pKa = 5.62EE117 pKa = 5.34YY118 pKa = 11.64DD119 pKa = 5.39DD120 pKa = 6.78DD121 pKa = 6.78DD122 pKa = 5.86DD123 pKa = 6.48LPLEE127 pKa = 3.97QLLGGGSKK135 pKa = 10.57RR136 pKa = 11.84SDD138 pKa = 3.24NTSQKK143 pKa = 10.49YY144 pKa = 10.43LIYY147 pKa = 10.44QLDD150 pKa = 3.44KK151 pKa = 10.87LYY153 pKa = 9.91PSKK156 pKa = 11.07VFEE159 pKa = 4.22PEE161 pKa = 3.34KK162 pKa = 10.3LQRR165 pKa = 11.84EE166 pKa = 4.25NMTEE170 pKa = 3.88QFEE173 pKa = 4.56DD174 pKa = 4.14DD175 pKa = 4.12SLEE178 pKa = 4.17LAQLYY183 pKa = 10.35YY184 pKa = 10.83DD185 pKa = 3.96YY186 pKa = 11.63LNASEE191 pKa = 5.01NGYY194 pKa = 8.58SEE196 pKa = 5.04CEE198 pKa = 3.74ARR200 pKa = 11.84QVAGLEE206 pKa = 3.73KK207 pKa = 10.92DD208 pKa = 3.52LFFRR212 pKa = 11.84MARR215 pKa = 11.84KK216 pKa = 8.92AADD219 pKa = 3.13LWSS222 pKa = 3.58
Molecular weight: 25.87 kDa
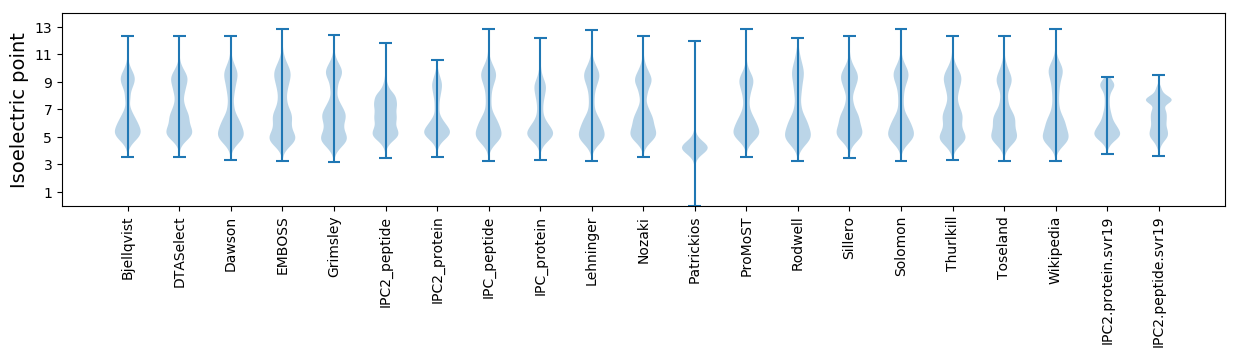
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1N6TCB9|A0A1N6TCB9_9BACT Cation/acetate symporter OS=Pontibacter lucknowensis OX=1077936 GN=SAMN05421545_0240 PE=3 SV=1
MM1 pKa = 7.25KK2 pKa = 10.08RR3 pKa = 11.84YY4 pKa = 9.58RR5 pKa = 11.84SWIGGIVLLGAMSCTSLSEE24 pKa = 4.26GGRR27 pKa = 11.84GTLPPDD33 pKa = 4.05PGHH36 pKa = 4.94TQRR39 pKa = 11.84VRR41 pKa = 11.84VSEE44 pKa = 4.14GDD46 pKa = 3.36PVRR49 pKa = 11.84RR50 pKa = 11.84QMYY53 pKa = 8.77EE54 pKa = 3.5QASDD58 pKa = 3.08INRR61 pKa = 11.84KK62 pKa = 9.57RR63 pKa = 11.84NIEE66 pKa = 4.08EE67 pKa = 3.85YY68 pKa = 10.76DD69 pKa = 3.26RR70 pKa = 11.84NAPVNLPPEE79 pKa = 4.27VRR81 pKa = 11.84PLQLPHH87 pKa = 7.2APVDD91 pKa = 4.08STRR94 pKa = 11.84HH95 pKa = 4.45RR96 pKa = 11.84VLRR99 pKa = 11.84ILPTRR104 pKa = 11.84PPVGNNNN111 pKa = 3.22
MM1 pKa = 7.25KK2 pKa = 10.08RR3 pKa = 11.84YY4 pKa = 9.58RR5 pKa = 11.84SWIGGIVLLGAMSCTSLSEE24 pKa = 4.26GGRR27 pKa = 11.84GTLPPDD33 pKa = 4.05PGHH36 pKa = 4.94TQRR39 pKa = 11.84VRR41 pKa = 11.84VSEE44 pKa = 4.14GDD46 pKa = 3.36PVRR49 pKa = 11.84RR50 pKa = 11.84QMYY53 pKa = 8.77EE54 pKa = 3.5QASDD58 pKa = 3.08INRR61 pKa = 11.84KK62 pKa = 9.57RR63 pKa = 11.84NIEE66 pKa = 4.08EE67 pKa = 3.85YY68 pKa = 10.76DD69 pKa = 3.26RR70 pKa = 11.84NAPVNLPPEE79 pKa = 4.27VRR81 pKa = 11.84PLQLPHH87 pKa = 7.2APVDD91 pKa = 4.08STRR94 pKa = 11.84HH95 pKa = 4.45RR96 pKa = 11.84VLRR99 pKa = 11.84ILPTRR104 pKa = 11.84PPVGNNNN111 pKa = 3.22
Molecular weight: 12.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1318896 |
29 |
3822 |
338.1 |
37.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.02 ± 0.036 | 0.701 ± 0.013 |
5.177 ± 0.029 | 6.515 ± 0.042 |
4.503 ± 0.029 | 7.001 ± 0.04 |
2.06 ± 0.024 | 6.157 ± 0.039 |
5.745 ± 0.04 | 10.02 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.524 ± 0.021 | 4.623 ± 0.034 |
4.104 ± 0.024 | 4.308 ± 0.03 |
4.909 ± 0.035 | 5.955 ± 0.034 |
5.746 ± 0.049 | 6.868 ± 0.031 |
1.101 ± 0.014 | 3.962 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
