
Halorientalis sp. IM1011
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Halobacteriales; Haloarculaceae; Halorientalis; unclassified Halorientalis
Average proteome isoelectric point is 4.82
Get precalculated fractions of proteins
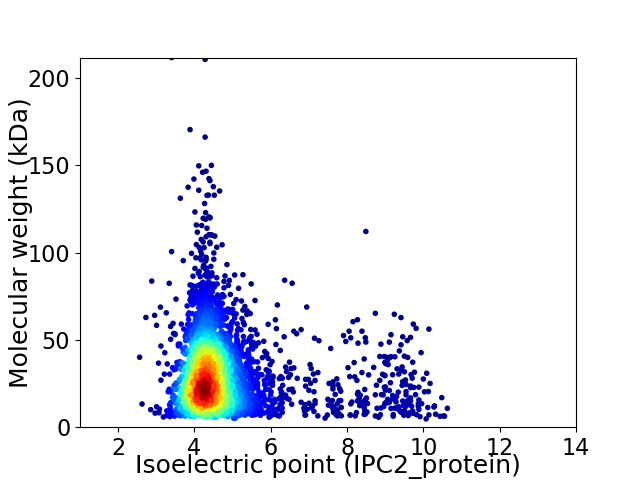
Virtual 2D-PAGE plot for 3726 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Q1FEM9|A0A1Q1FEM9_9EURY Uncharacterized protein OS=Halorientalis sp. IM1011 OX=1932360 GN=BV210_00405 PE=4 SV=1
MM1 pKa = 7.44EE2 pKa = 4.82LTWHH6 pKa = 6.53GHH8 pKa = 4.65ATWHH12 pKa = 5.88VSVGDD17 pKa = 3.67TDD19 pKa = 4.19ILIDD23 pKa = 4.39PFFDD27 pKa = 3.85NPKK30 pKa = 9.1TDD32 pKa = 3.84AEE34 pKa = 4.73AGDD37 pKa = 4.23FDD39 pKa = 5.64PDD41 pKa = 3.76VVLVTHH47 pKa = 6.35GHH49 pKa = 7.05DD50 pKa = 3.45DD51 pKa = 3.36HH52 pKa = 8.2VADD55 pKa = 4.5VGAFTDD61 pKa = 3.58ATLAGTPEE69 pKa = 4.46VVGHH73 pKa = 6.79LSGEE77 pKa = 4.0HH78 pKa = 7.1DD79 pKa = 3.72FAEE82 pKa = 4.71DD83 pKa = 3.69DD84 pKa = 3.9VVGFNLGGTVEE95 pKa = 4.88FGDD98 pKa = 4.82AYY100 pKa = 10.38VTMHH104 pKa = 7.24RR105 pKa = 11.84ADD107 pKa = 3.49HH108 pKa = 5.83TNGMGEE114 pKa = 4.35GFVAPSGGNCAGYY127 pKa = 10.31VISDD131 pKa = 3.79TVPTQIEE138 pKa = 4.54DD139 pKa = 3.52EE140 pKa = 4.67DD141 pKa = 4.11STTFYY146 pKa = 10.83HH147 pKa = 7.38AGDD150 pKa = 3.75TGLMTEE156 pKa = 4.06MRR158 pKa = 11.84EE159 pKa = 4.24VIGPYY164 pKa = 10.13LEE166 pKa = 4.97PDD168 pKa = 3.24AAALPVGDD176 pKa = 4.97HH177 pKa = 5.53FTMGPMQAAIAVDD190 pKa = 3.74WLDD193 pKa = 3.34VDD195 pKa = 4.28HH196 pKa = 7.44AFPMHH201 pKa = 7.05YY202 pKa = 8.85DD203 pKa = 3.81TFPPIEE209 pKa = 4.31MDD211 pKa = 3.24PDD213 pKa = 3.65RR214 pKa = 11.84FVDD217 pKa = 4.4EE218 pKa = 4.48VAATGSDD225 pKa = 3.64AEE227 pKa = 4.36VHH229 pKa = 6.26VLDD232 pKa = 4.85GDD234 pKa = 3.74EE235 pKa = 4.97SFDD238 pKa = 4.76LGTT241 pKa = 4.14
MM1 pKa = 7.44EE2 pKa = 4.82LTWHH6 pKa = 6.53GHH8 pKa = 4.65ATWHH12 pKa = 5.88VSVGDD17 pKa = 3.67TDD19 pKa = 4.19ILIDD23 pKa = 4.39PFFDD27 pKa = 3.85NPKK30 pKa = 9.1TDD32 pKa = 3.84AEE34 pKa = 4.73AGDD37 pKa = 4.23FDD39 pKa = 5.64PDD41 pKa = 3.76VVLVTHH47 pKa = 6.35GHH49 pKa = 7.05DD50 pKa = 3.45DD51 pKa = 3.36HH52 pKa = 8.2VADD55 pKa = 4.5VGAFTDD61 pKa = 3.58ATLAGTPEE69 pKa = 4.46VVGHH73 pKa = 6.79LSGEE77 pKa = 4.0HH78 pKa = 7.1DD79 pKa = 3.72FAEE82 pKa = 4.71DD83 pKa = 3.69DD84 pKa = 3.9VVGFNLGGTVEE95 pKa = 4.88FGDD98 pKa = 4.82AYY100 pKa = 10.38VTMHH104 pKa = 7.24RR105 pKa = 11.84ADD107 pKa = 3.49HH108 pKa = 5.83TNGMGEE114 pKa = 4.35GFVAPSGGNCAGYY127 pKa = 10.31VISDD131 pKa = 3.79TVPTQIEE138 pKa = 4.54DD139 pKa = 3.52EE140 pKa = 4.67DD141 pKa = 4.11STTFYY146 pKa = 10.83HH147 pKa = 7.38AGDD150 pKa = 3.75TGLMTEE156 pKa = 4.06MRR158 pKa = 11.84EE159 pKa = 4.24VIGPYY164 pKa = 10.13LEE166 pKa = 4.97PDD168 pKa = 3.24AAALPVGDD176 pKa = 4.97HH177 pKa = 5.53FTMGPMQAAIAVDD190 pKa = 3.74WLDD193 pKa = 3.34VDD195 pKa = 4.28HH196 pKa = 7.44AFPMHH201 pKa = 7.05YY202 pKa = 8.85DD203 pKa = 3.81TFPPIEE209 pKa = 4.31MDD211 pKa = 3.24PDD213 pKa = 3.65RR214 pKa = 11.84FVDD217 pKa = 4.4EE218 pKa = 4.48VAATGSDD225 pKa = 3.64AEE227 pKa = 4.36VHH229 pKa = 6.26VLDD232 pKa = 4.85GDD234 pKa = 3.74EE235 pKa = 4.97SFDD238 pKa = 4.76LGTT241 pKa = 4.14
Molecular weight: 25.82 kDa
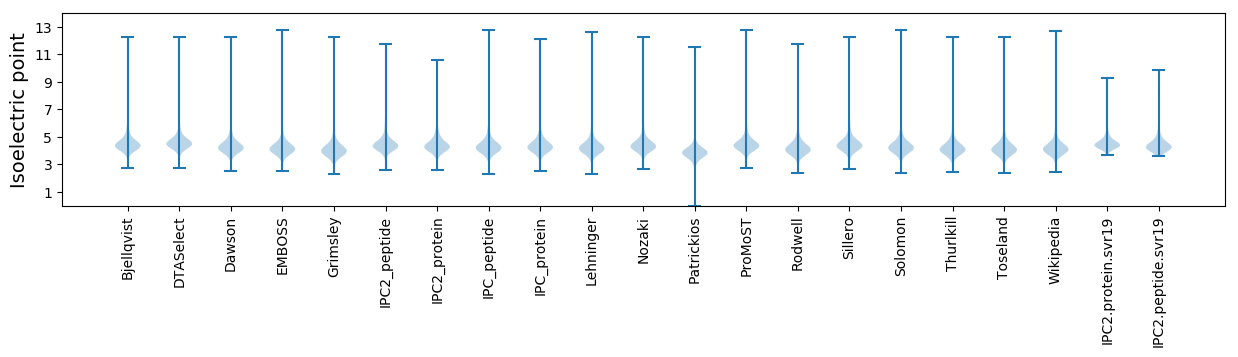
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Q1FHR6|A0A1Q1FHR6_9EURY MBL fold metallo-hydrolase OS=Halorientalis sp. IM1011 OX=1932360 GN=BV210_06225 PE=4 SV=1
MM1 pKa = 7.02TRR3 pKa = 11.84SFYY6 pKa = 10.88SHH8 pKa = 6.79IRR10 pKa = 11.84EE11 pKa = 4.12AWEE14 pKa = 5.12DD15 pKa = 3.61PDD17 pKa = 5.76DD18 pKa = 4.4GQLAEE23 pKa = 4.85LQWQRR28 pKa = 11.84QQEE31 pKa = 4.13WRR33 pKa = 11.84DD34 pKa = 3.31QGALEE39 pKa = 4.11RR40 pKa = 11.84VEE42 pKa = 4.47RR43 pKa = 11.84PTRR46 pKa = 11.84LDD48 pKa = 3.0KK49 pKa = 11.23ARR51 pKa = 11.84SLGYY55 pKa = 9.5KK56 pKa = 9.39AKK58 pKa = 10.36QGIVVVRR65 pKa = 11.84ASIRR69 pKa = 11.84KK70 pKa = 9.0GGARR74 pKa = 11.84KK75 pKa = 9.23QRR77 pKa = 11.84FTAGRR82 pKa = 11.84RR83 pKa = 11.84SKK85 pKa = 10.54RR86 pKa = 11.84QGVTRR91 pKa = 11.84ITRR94 pKa = 11.84RR95 pKa = 11.84KK96 pKa = 8.26NLQRR100 pKa = 11.84VAEE103 pKa = 4.12EE104 pKa = 3.75RR105 pKa = 11.84ATRR108 pKa = 11.84KK109 pKa = 9.49YY110 pKa = 10.94RR111 pKa = 11.84NLRR114 pKa = 11.84VLNSYY119 pKa = 10.83SVGQDD124 pKa = 3.09GRR126 pKa = 11.84QKK128 pKa = 8.5WFEE131 pKa = 4.19VILVDD136 pKa = 3.49PHH138 pKa = 7.55HH139 pKa = 6.74PAIEE143 pKa = 4.25NDD145 pKa = 4.51DD146 pKa = 4.12DD147 pKa = 5.67LNWICDD153 pKa = 3.4DD154 pKa = 3.81SQRR157 pKa = 11.84GRR159 pKa = 11.84AYY161 pKa = 10.59RR162 pKa = 11.84GLTNAGQSNRR172 pKa = 11.84GLDD175 pKa = 3.43QKK177 pKa = 11.66GKK179 pKa = 8.43GTEE182 pKa = 3.99HH183 pKa = 6.37NRR185 pKa = 11.84PSNNSGRR192 pKa = 11.84GRR194 pKa = 11.84AKK196 pKa = 10.54
MM1 pKa = 7.02TRR3 pKa = 11.84SFYY6 pKa = 10.88SHH8 pKa = 6.79IRR10 pKa = 11.84EE11 pKa = 4.12AWEE14 pKa = 5.12DD15 pKa = 3.61PDD17 pKa = 5.76DD18 pKa = 4.4GQLAEE23 pKa = 4.85LQWQRR28 pKa = 11.84QQEE31 pKa = 4.13WRR33 pKa = 11.84DD34 pKa = 3.31QGALEE39 pKa = 4.11RR40 pKa = 11.84VEE42 pKa = 4.47RR43 pKa = 11.84PTRR46 pKa = 11.84LDD48 pKa = 3.0KK49 pKa = 11.23ARR51 pKa = 11.84SLGYY55 pKa = 9.5KK56 pKa = 9.39AKK58 pKa = 10.36QGIVVVRR65 pKa = 11.84ASIRR69 pKa = 11.84KK70 pKa = 9.0GGARR74 pKa = 11.84KK75 pKa = 9.23QRR77 pKa = 11.84FTAGRR82 pKa = 11.84RR83 pKa = 11.84SKK85 pKa = 10.54RR86 pKa = 11.84QGVTRR91 pKa = 11.84ITRR94 pKa = 11.84RR95 pKa = 11.84KK96 pKa = 8.26NLQRR100 pKa = 11.84VAEE103 pKa = 4.12EE104 pKa = 3.75RR105 pKa = 11.84ATRR108 pKa = 11.84KK109 pKa = 9.49YY110 pKa = 10.94RR111 pKa = 11.84NLRR114 pKa = 11.84VLNSYY119 pKa = 10.83SVGQDD124 pKa = 3.09GRR126 pKa = 11.84QKK128 pKa = 8.5WFEE131 pKa = 4.19VILVDD136 pKa = 3.49PHH138 pKa = 7.55HH139 pKa = 6.74PAIEE143 pKa = 4.25NDD145 pKa = 4.51DD146 pKa = 4.12DD147 pKa = 5.67LNWICDD153 pKa = 3.4DD154 pKa = 3.81SQRR157 pKa = 11.84GRR159 pKa = 11.84AYY161 pKa = 10.59RR162 pKa = 11.84GLTNAGQSNRR172 pKa = 11.84GLDD175 pKa = 3.43QKK177 pKa = 11.66GKK179 pKa = 8.43GTEE182 pKa = 3.99HH183 pKa = 6.37NRR185 pKa = 11.84PSNNSGRR192 pKa = 11.84GRR194 pKa = 11.84AKK196 pKa = 10.54
Molecular weight: 22.76 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1076951 |
39 |
2026 |
289.0 |
31.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.766 ± 0.06 | 0.702 ± 0.011 |
8.691 ± 0.052 | 8.671 ± 0.064 |
3.3 ± 0.027 | 8.57 ± 0.038 |
1.953 ± 0.018 | 3.974 ± 0.034 |
1.765 ± 0.024 | 8.93 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.772 ± 0.019 | 2.27 ± 0.024 |
4.708 ± 0.024 | 2.581 ± 0.027 |
6.591 ± 0.046 | 5.3 ± 0.035 |
6.571 ± 0.038 | 9.103 ± 0.045 |
1.131 ± 0.015 | 2.651 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
