
Citrus leaf blotch virus (isolate Nagami kumquat/France/SRA-153/1984) (CLBV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Tymovirales; Betaflexiviridae; Trivirinae; Citrivirus; Citrus leaf blotch virus
Average proteome isoelectric point is 6.05
Get precalculated fractions of proteins
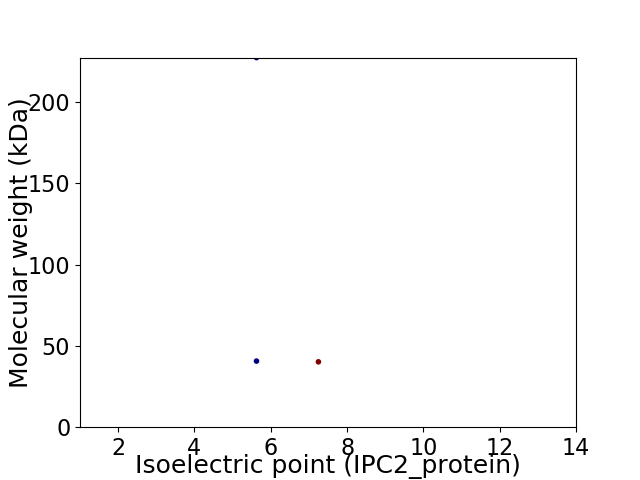
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
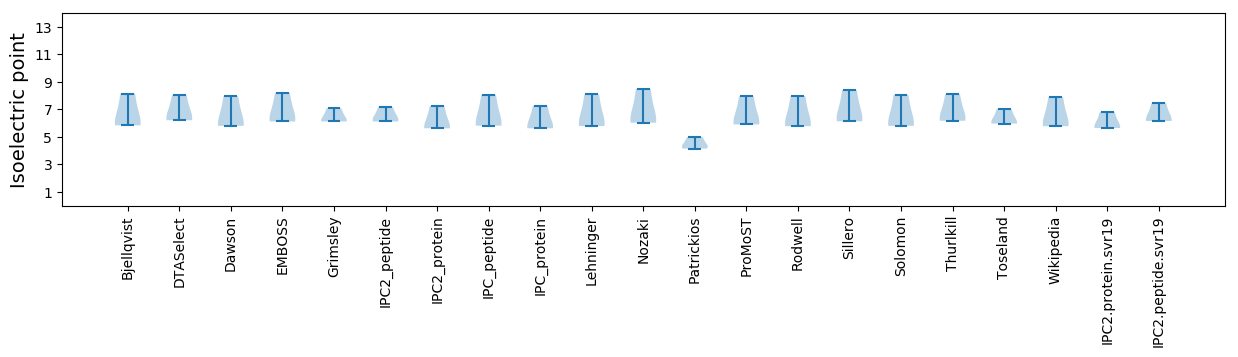
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q91QZ2|MOVP_CLBVS Putative movement protein OS=Citrus leaf blotch virus (isolate Nagami kumquat/France/SRA-153/1984) OX=686981 GN=ORF2 PE=4 SV=1
MM1 pKa = 8.02KK2 pKa = 9.34ITNDD6 pKa = 2.86NAATINYY13 pKa = 7.34WLAIVEE19 pKa = 4.31PFLTSDD25 pKa = 3.5EE26 pKa = 4.84DD27 pKa = 4.18RR28 pKa = 11.84NSDD31 pKa = 3.91DD32 pKa = 4.06IIRR35 pKa = 11.84KK36 pKa = 8.45FRR38 pKa = 11.84AVVAEE43 pKa = 4.29HH44 pKa = 7.19GDD46 pKa = 3.71TEE48 pKa = 4.43EE49 pKa = 3.9VDD51 pKa = 3.62PEE53 pKa = 4.24VFFAIFSILATKK65 pKa = 9.77YY66 pKa = 10.14GRR68 pKa = 11.84VYY70 pKa = 10.32SKK72 pKa = 10.66KK73 pKa = 10.03VEE75 pKa = 4.09EE76 pKa = 4.44LNEE79 pKa = 4.11SLKK82 pKa = 11.07AAILAGAEE90 pKa = 4.29AEE92 pKa = 4.18DD93 pKa = 4.97LRR95 pKa = 11.84NKK97 pKa = 10.56LKK99 pKa = 10.77DD100 pKa = 2.93ISQRR104 pKa = 11.84YY105 pKa = 8.57ASQLEE110 pKa = 4.0ITADD114 pKa = 3.63RR115 pKa = 11.84EE116 pKa = 4.16QQLEE120 pKa = 4.1SLKK123 pKa = 10.93KK124 pKa = 10.0KK125 pKa = 10.05GHH127 pKa = 5.38EE128 pKa = 4.05QPLTGSGSSEE138 pKa = 3.79PVHH141 pKa = 7.01AEE143 pKa = 3.89SAHH146 pKa = 5.87APQLHH151 pKa = 5.65VVNDD155 pKa = 3.9LQQFYY160 pKa = 10.66IPFNEE165 pKa = 4.24YY166 pKa = 10.05PSLTQSIGTSDD177 pKa = 3.28IANDD181 pKa = 3.35EE182 pKa = 4.11HH183 pKa = 7.2LKK185 pKa = 10.51RR186 pKa = 11.84VQLTLKK192 pKa = 9.17ITDD195 pKa = 3.52TKK197 pKa = 10.79VFSRR201 pKa = 11.84TGFEE205 pKa = 4.12FAISCGSRR213 pKa = 11.84STSDD217 pKa = 3.05KK218 pKa = 11.14DD219 pKa = 3.56PYY221 pKa = 11.19DD222 pKa = 4.47GIIKK226 pKa = 10.22ISGKK230 pKa = 7.87SHH232 pKa = 4.67MRR234 pKa = 11.84KK235 pKa = 10.06DD236 pKa = 2.68IAYY239 pKa = 9.37AIRR242 pKa = 11.84TSGITVRR249 pKa = 11.84QFCAAFANLYY259 pKa = 9.97WNFNLARR266 pKa = 11.84NTPPEE271 pKa = 3.7NWRR274 pKa = 11.84KK275 pKa = 10.23KK276 pKa = 10.68GFTEE280 pKa = 4.3GTKK283 pKa = 10.17FAAFDD288 pKa = 3.29FFYY291 pKa = 10.95AVGSNAAIPTEE302 pKa = 4.02ADD304 pKa = 3.11GSVRR308 pKa = 11.84LIRR311 pKa = 11.84PPTNEE316 pKa = 3.61EE317 pKa = 3.85NEE319 pKa = 4.26ANSAMRR325 pKa = 11.84YY326 pKa = 9.39ADD328 pKa = 3.54IYY330 pKa = 9.42EE331 pKa = 4.36QNSKK335 pKa = 7.91TAGHH339 pKa = 5.6VTSSPLYY346 pKa = 9.81NRR348 pKa = 11.84GSSYY352 pKa = 10.54EE353 pKa = 4.22SKK355 pKa = 11.12NKK357 pKa = 10.48AKK359 pKa = 10.46LLEE362 pKa = 4.16MM363 pKa = 5.41
MM1 pKa = 8.02KK2 pKa = 9.34ITNDD6 pKa = 2.86NAATINYY13 pKa = 7.34WLAIVEE19 pKa = 4.31PFLTSDD25 pKa = 3.5EE26 pKa = 4.84DD27 pKa = 4.18RR28 pKa = 11.84NSDD31 pKa = 3.91DD32 pKa = 4.06IIRR35 pKa = 11.84KK36 pKa = 8.45FRR38 pKa = 11.84AVVAEE43 pKa = 4.29HH44 pKa = 7.19GDD46 pKa = 3.71TEE48 pKa = 4.43EE49 pKa = 3.9VDD51 pKa = 3.62PEE53 pKa = 4.24VFFAIFSILATKK65 pKa = 9.77YY66 pKa = 10.14GRR68 pKa = 11.84VYY70 pKa = 10.32SKK72 pKa = 10.66KK73 pKa = 10.03VEE75 pKa = 4.09EE76 pKa = 4.44LNEE79 pKa = 4.11SLKK82 pKa = 11.07AAILAGAEE90 pKa = 4.29AEE92 pKa = 4.18DD93 pKa = 4.97LRR95 pKa = 11.84NKK97 pKa = 10.56LKK99 pKa = 10.77DD100 pKa = 2.93ISQRR104 pKa = 11.84YY105 pKa = 8.57ASQLEE110 pKa = 4.0ITADD114 pKa = 3.63RR115 pKa = 11.84EE116 pKa = 4.16QQLEE120 pKa = 4.1SLKK123 pKa = 10.93KK124 pKa = 10.0KK125 pKa = 10.05GHH127 pKa = 5.38EE128 pKa = 4.05QPLTGSGSSEE138 pKa = 3.79PVHH141 pKa = 7.01AEE143 pKa = 3.89SAHH146 pKa = 5.87APQLHH151 pKa = 5.65VVNDD155 pKa = 3.9LQQFYY160 pKa = 10.66IPFNEE165 pKa = 4.24YY166 pKa = 10.05PSLTQSIGTSDD177 pKa = 3.28IANDD181 pKa = 3.35EE182 pKa = 4.11HH183 pKa = 7.2LKK185 pKa = 10.51RR186 pKa = 11.84VQLTLKK192 pKa = 9.17ITDD195 pKa = 3.52TKK197 pKa = 10.79VFSRR201 pKa = 11.84TGFEE205 pKa = 4.12FAISCGSRR213 pKa = 11.84STSDD217 pKa = 3.05KK218 pKa = 11.14DD219 pKa = 3.56PYY221 pKa = 11.19DD222 pKa = 4.47GIIKK226 pKa = 10.22ISGKK230 pKa = 7.87SHH232 pKa = 4.67MRR234 pKa = 11.84KK235 pKa = 10.06DD236 pKa = 2.68IAYY239 pKa = 9.37AIRR242 pKa = 11.84TSGITVRR249 pKa = 11.84QFCAAFANLYY259 pKa = 9.97WNFNLARR266 pKa = 11.84NTPPEE271 pKa = 3.7NWRR274 pKa = 11.84KK275 pKa = 10.23KK276 pKa = 10.68GFTEE280 pKa = 4.3GTKK283 pKa = 10.17FAAFDD288 pKa = 3.29FFYY291 pKa = 10.95AVGSNAAIPTEE302 pKa = 4.02ADD304 pKa = 3.11GSVRR308 pKa = 11.84LIRR311 pKa = 11.84PPTNEE316 pKa = 3.61EE317 pKa = 3.85NEE319 pKa = 4.26ANSAMRR325 pKa = 11.84YY326 pKa = 9.39ADD328 pKa = 3.54IYY330 pKa = 9.42EE331 pKa = 4.36QNSKK335 pKa = 7.91TAGHH339 pKa = 5.6VTSSPLYY346 pKa = 9.81NRR348 pKa = 11.84GSSYY352 pKa = 10.54EE353 pKa = 4.22SKK355 pKa = 11.12NKK357 pKa = 10.48AKK359 pKa = 10.46LLEE362 pKa = 4.16MM363 pKa = 5.41
Molecular weight: 40.68 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q91QZ3|RDRP_CLBVS RNA replication polyprotein OS=Citrus leaf blotch virus (isolate Nagami kumquat/France/SRA-153/1984) OX=686981 GN=ORF1 PE=3 SV=1
MM1 pKa = 7.73ASLINVSSLVNRR13 pKa = 11.84VKK15 pKa = 10.75LDD17 pKa = 3.31QSIIGSDD24 pKa = 4.49EE25 pKa = 3.75INKK28 pKa = 10.14LYY30 pKa = 10.95GSDD33 pKa = 3.46APLVFKK39 pKa = 11.12DD40 pKa = 3.84EE41 pKa = 4.32VKK43 pKa = 10.33MVIPGNAEE51 pKa = 3.48GEE53 pKa = 4.38AIKK56 pKa = 10.63LQANILTADD65 pKa = 4.11RR66 pKa = 11.84LQSIRR71 pKa = 11.84NAKK74 pKa = 10.0VNGKK78 pKa = 8.79EE79 pKa = 3.54AAYY82 pKa = 10.06LHH84 pKa = 6.59LGFVPIAIRR93 pKa = 11.84SLLPSGNEE101 pKa = 4.1QIWGRR106 pKa = 11.84CALVDD111 pKa = 3.58TSRR114 pKa = 11.84TRR116 pKa = 11.84AEE118 pKa = 3.81TAVIDD123 pKa = 3.71EE124 pKa = 4.96FEE126 pKa = 4.31FKK128 pKa = 9.35FTKK131 pKa = 9.95KK132 pKa = 10.36QPFAAKK138 pKa = 10.46LLTINAAVDD147 pKa = 3.55INCKK151 pKa = 10.2VSVGSIQVLLEE162 pKa = 3.67LHH164 pKa = 6.49GVDD167 pKa = 4.03LRR169 pKa = 11.84EE170 pKa = 4.1EE171 pKa = 4.13RR172 pKa = 11.84SVAAIITGLTCTPTNKK188 pKa = 9.03MVLLHH193 pKa = 6.93KK194 pKa = 10.24IEE196 pKa = 5.15CDD198 pKa = 3.3TPKK201 pKa = 10.31WSLCNIIEE209 pKa = 4.14QVEE212 pKa = 4.12DD213 pKa = 3.57EE214 pKa = 4.71EE215 pKa = 4.35EE216 pKa = 4.13SKK218 pKa = 11.0KK219 pKa = 10.96AFEE222 pKa = 4.39NMFNASSSNLIDD234 pKa = 4.7LGQEE238 pKa = 3.4QWLDD242 pKa = 3.33EE243 pKa = 4.5GKK245 pKa = 8.52RR246 pKa = 11.84TPLIGSLAIKK256 pKa = 10.58GFGRR260 pKa = 11.84KK261 pKa = 8.29VMPVRR266 pKa = 11.84RR267 pKa = 11.84RR268 pKa = 11.84NLTTRR273 pKa = 11.84NLMKK277 pKa = 10.48DD278 pKa = 3.5YY279 pKa = 10.51VSHH282 pKa = 6.28VKK284 pKa = 10.56SEE286 pKa = 4.06TASLKK291 pKa = 10.18RR292 pKa = 11.84SQSGRR297 pKa = 11.84DD298 pKa = 3.01WGNDD302 pKa = 2.93RR303 pKa = 11.84LRR305 pKa = 11.84KK306 pKa = 9.57YY307 pKa = 10.79LEE309 pKa = 4.07EE310 pKa = 3.98QALEE314 pKa = 4.11QARR317 pKa = 11.84SSTDD321 pKa = 3.06HH322 pKa = 6.46QLVKK326 pKa = 10.69APKK329 pKa = 8.91FKK331 pKa = 10.29TIEE334 pKa = 3.98VTGLEE339 pKa = 4.43TVHH342 pKa = 6.93DD343 pKa = 4.88LKK345 pKa = 11.5DD346 pKa = 4.28KK347 pKa = 10.53IDD349 pKa = 3.65KK350 pKa = 10.89AEE352 pKa = 4.15SSTASDD358 pKa = 3.44TGTKK362 pKa = 9.97
MM1 pKa = 7.73ASLINVSSLVNRR13 pKa = 11.84VKK15 pKa = 10.75LDD17 pKa = 3.31QSIIGSDD24 pKa = 4.49EE25 pKa = 3.75INKK28 pKa = 10.14LYY30 pKa = 10.95GSDD33 pKa = 3.46APLVFKK39 pKa = 11.12DD40 pKa = 3.84EE41 pKa = 4.32VKK43 pKa = 10.33MVIPGNAEE51 pKa = 3.48GEE53 pKa = 4.38AIKK56 pKa = 10.63LQANILTADD65 pKa = 4.11RR66 pKa = 11.84LQSIRR71 pKa = 11.84NAKK74 pKa = 10.0VNGKK78 pKa = 8.79EE79 pKa = 3.54AAYY82 pKa = 10.06LHH84 pKa = 6.59LGFVPIAIRR93 pKa = 11.84SLLPSGNEE101 pKa = 4.1QIWGRR106 pKa = 11.84CALVDD111 pKa = 3.58TSRR114 pKa = 11.84TRR116 pKa = 11.84AEE118 pKa = 3.81TAVIDD123 pKa = 3.71EE124 pKa = 4.96FEE126 pKa = 4.31FKK128 pKa = 9.35FTKK131 pKa = 9.95KK132 pKa = 10.36QPFAAKK138 pKa = 10.46LLTINAAVDD147 pKa = 3.55INCKK151 pKa = 10.2VSVGSIQVLLEE162 pKa = 3.67LHH164 pKa = 6.49GVDD167 pKa = 4.03LRR169 pKa = 11.84EE170 pKa = 4.1EE171 pKa = 4.13RR172 pKa = 11.84SVAAIITGLTCTPTNKK188 pKa = 9.03MVLLHH193 pKa = 6.93KK194 pKa = 10.24IEE196 pKa = 5.15CDD198 pKa = 3.3TPKK201 pKa = 10.31WSLCNIIEE209 pKa = 4.14QVEE212 pKa = 4.12DD213 pKa = 3.57EE214 pKa = 4.71EE215 pKa = 4.35EE216 pKa = 4.13SKK218 pKa = 11.0KK219 pKa = 10.96AFEE222 pKa = 4.39NMFNASSSNLIDD234 pKa = 4.7LGQEE238 pKa = 3.4QWLDD242 pKa = 3.33EE243 pKa = 4.5GKK245 pKa = 8.52RR246 pKa = 11.84TPLIGSLAIKK256 pKa = 10.58GFGRR260 pKa = 11.84KK261 pKa = 8.29VMPVRR266 pKa = 11.84RR267 pKa = 11.84RR268 pKa = 11.84NLTTRR273 pKa = 11.84NLMKK277 pKa = 10.48DD278 pKa = 3.5YY279 pKa = 10.51VSHH282 pKa = 6.28VKK284 pKa = 10.56SEE286 pKa = 4.06TASLKK291 pKa = 10.18RR292 pKa = 11.84SQSGRR297 pKa = 11.84DD298 pKa = 3.01WGNDD302 pKa = 2.93RR303 pKa = 11.84LRR305 pKa = 11.84KK306 pKa = 9.57YY307 pKa = 10.79LEE309 pKa = 4.07EE310 pKa = 3.98QALEE314 pKa = 4.11QARR317 pKa = 11.84SSTDD321 pKa = 3.06HH322 pKa = 6.46QLVKK326 pKa = 10.69APKK329 pKa = 8.91FKK331 pKa = 10.29TIEE334 pKa = 3.98VTGLEE339 pKa = 4.43TVHH342 pKa = 6.93DD343 pKa = 4.88LKK345 pKa = 11.5DD346 pKa = 4.28KK347 pKa = 10.53IDD349 pKa = 3.65KK350 pKa = 10.89AEE352 pKa = 4.15SSTASDD358 pKa = 3.44TGTKK362 pKa = 9.97
Molecular weight: 40.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2687 |
362 |
1962 |
895.7 |
102.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.215 ± 1.539 | 2.01 ± 0.613 |
5.769 ± 0.014 | 8.336 ± 0.26 |
6.699 ± 1.564 | 4.987 ± 0.158 |
2.642 ± 0.392 | 6.401 ± 0.197 |
7.146 ± 0.395 | 9.155 ± 0.772 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.861 ± 0.294 | 5.359 ± 0.137 |
3.052 ± 0.246 | 3.089 ± 0.198 |
5.434 ± 0.06 | 7.294 ± 0.652 |
4.801 ± 0.786 | 5.62 ± 0.413 |
1.042 ± 0.067 | 3.089 ± 0.529 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
