
Rickettsiales bacterium Ac37b
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rickettsiales; unclassified Rickettsiales
Average proteome isoelectric point is 6.87
Get precalculated fractions of proteins
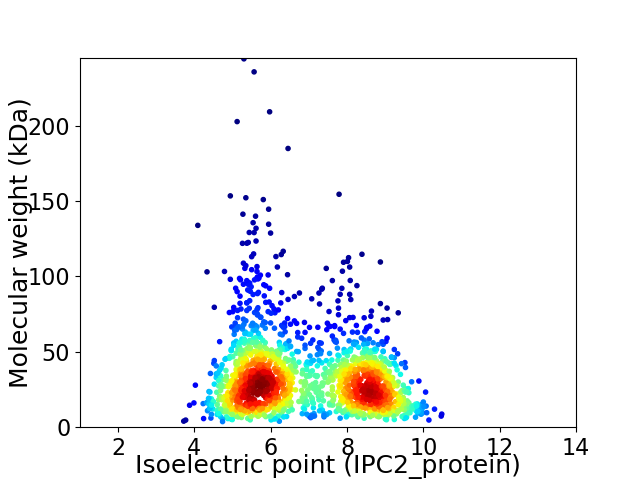
Virtual 2D-PAGE plot for 1659 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
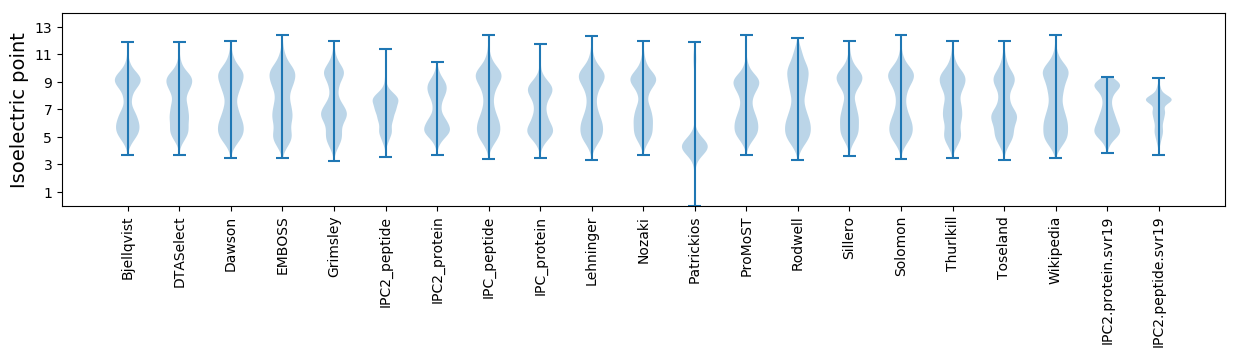
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A077FIX2|A0A077FIX2_9RICK Uncharacterized protein OS=Rickettsiales bacterium Ac37b OX=1528098 GN=NOVO_05465 PE=4 SV=1
MM1 pKa = 7.16IAVMTYY7 pKa = 10.04LKK9 pKa = 10.15WLFYY13 pKa = 10.29PILALFITQPTFAAPIWTTSFGSQGTGDD41 pKa = 3.81GQFDD45 pKa = 4.33NPTGITTLPTGEE57 pKa = 4.37IYY59 pKa = 11.06VVDD62 pKa = 4.3GNRR65 pKa = 11.84NDD67 pKa = 3.57VQIFNPDD74 pKa = 3.0TTFSSKK80 pKa = 10.42FGSPGNGNGEE90 pKa = 4.21FNLPISIFRR99 pKa = 11.84SPSTGLLYY107 pKa = 10.45ISDD110 pKa = 4.12NFNHH114 pKa = 6.61RR115 pKa = 11.84VQIFNPDD122 pKa = 3.03GTYY125 pKa = 10.38SAQFGSFGDD134 pKa = 3.82GDD136 pKa = 4.42GQFMDD141 pKa = 4.48HH142 pKa = 6.71YY143 pKa = 9.75GITMDD148 pKa = 3.59TVNNKK153 pKa = 9.29FFVSDD158 pKa = 3.31VGNNNNPRR166 pKa = 11.84VQIFNDD172 pKa = 3.32DD173 pKa = 3.19GTYY176 pKa = 10.3FSQFGGFGTANGEE189 pKa = 4.43FYY191 pKa = 11.31SPMSLTFGNSQLYY204 pKa = 8.56VADD207 pKa = 3.73YY208 pKa = 10.13FLNNIQTFDD217 pKa = 3.43EE218 pKa = 4.42NANFLNKK225 pKa = 9.68FDD227 pKa = 3.96VSSIPSVYY235 pKa = 10.49LSRR238 pKa = 11.84PRR240 pKa = 11.84DD241 pKa = 3.45LAVDD245 pKa = 3.88NAHH248 pKa = 7.37DD249 pKa = 3.78MYY251 pKa = 11.71
MM1 pKa = 7.16IAVMTYY7 pKa = 10.04LKK9 pKa = 10.15WLFYY13 pKa = 10.29PILALFITQPTFAAPIWTTSFGSQGTGDD41 pKa = 3.81GQFDD45 pKa = 4.33NPTGITTLPTGEE57 pKa = 4.37IYY59 pKa = 11.06VVDD62 pKa = 4.3GNRR65 pKa = 11.84NDD67 pKa = 3.57VQIFNPDD74 pKa = 3.0TTFSSKK80 pKa = 10.42FGSPGNGNGEE90 pKa = 4.21FNLPISIFRR99 pKa = 11.84SPSTGLLYY107 pKa = 10.45ISDD110 pKa = 4.12NFNHH114 pKa = 6.61RR115 pKa = 11.84VQIFNPDD122 pKa = 3.03GTYY125 pKa = 10.38SAQFGSFGDD134 pKa = 3.82GDD136 pKa = 4.42GQFMDD141 pKa = 4.48HH142 pKa = 6.71YY143 pKa = 9.75GITMDD148 pKa = 3.59TVNNKK153 pKa = 9.29FFVSDD158 pKa = 3.31VGNNNNPRR166 pKa = 11.84VQIFNDD172 pKa = 3.32DD173 pKa = 3.19GTYY176 pKa = 10.3FSQFGGFGTANGEE189 pKa = 4.43FYY191 pKa = 11.31SPMSLTFGNSQLYY204 pKa = 8.56VADD207 pKa = 3.73YY208 pKa = 10.13FLNNIQTFDD217 pKa = 3.43EE218 pKa = 4.42NANFLNKK225 pKa = 9.68FDD227 pKa = 3.96VSSIPSVYY235 pKa = 10.49LSRR238 pKa = 11.84PRR240 pKa = 11.84DD241 pKa = 3.45LAVDD245 pKa = 3.88NAHH248 pKa = 7.37DD249 pKa = 3.78MYY251 pKa = 11.71
Molecular weight: 27.88 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A077FGQ8|A0A077FGQ8_9RICK Thioredoxin OS=Rickettsiales bacterium Ac37b OX=1528098 GN=trxA PE=3 SV=1
MM1 pKa = 7.82NYY3 pKa = 10.14RR4 pKa = 11.84IPKK7 pKa = 8.62TLDD10 pKa = 2.99NPVRR14 pKa = 11.84CLGIPIDD21 pKa = 3.81ALIVFMGIWGGFVVFDD37 pKa = 4.35KK38 pKa = 11.25GLFGIPVGIVVSMLFGKK55 pKa = 10.06FRR57 pKa = 11.84SRR59 pKa = 11.84SIVRR63 pKa = 11.84RR64 pKa = 11.84FIRR67 pKa = 11.84FLYY70 pKa = 9.37WYY72 pKa = 9.48LPSEE76 pKa = 4.21MNFIKK81 pKa = 10.44GVQGHH86 pKa = 4.92QRR88 pKa = 11.84KK89 pKa = 9.41LICKK93 pKa = 9.32
MM1 pKa = 7.82NYY3 pKa = 10.14RR4 pKa = 11.84IPKK7 pKa = 8.62TLDD10 pKa = 2.99NPVRR14 pKa = 11.84CLGIPIDD21 pKa = 3.81ALIVFMGIWGGFVVFDD37 pKa = 4.35KK38 pKa = 11.25GLFGIPVGIVVSMLFGKK55 pKa = 10.06FRR57 pKa = 11.84SRR59 pKa = 11.84SIVRR63 pKa = 11.84RR64 pKa = 11.84FIRR67 pKa = 11.84FLYY70 pKa = 9.37WYY72 pKa = 9.48LPSEE76 pKa = 4.21MNFIKK81 pKa = 10.44GVQGHH86 pKa = 4.92QRR88 pKa = 11.84KK89 pKa = 9.41LICKK93 pKa = 9.32
Molecular weight: 10.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
520560 |
31 |
2243 |
313.8 |
35.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.289 ± 0.064 | 1.029 ± 0.023 |
4.878 ± 0.046 | 6.335 ± 0.067 |
4.199 ± 0.043 | 5.474 ± 0.067 |
2.168 ± 0.028 | 9.987 ± 0.072 |
7.732 ± 0.064 | 10.229 ± 0.073 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.261 ± 0.022 | 6.779 ± 0.067 |
3.317 ± 0.037 | 3.509 ± 0.037 |
3.409 ± 0.046 | 6.983 ± 0.045 |
5.19 ± 0.04 | 5.482 ± 0.046 |
0.805 ± 0.021 | 3.943 ± 0.045 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
