
Shahe sobemo-like virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.26
Get precalculated fractions of proteins
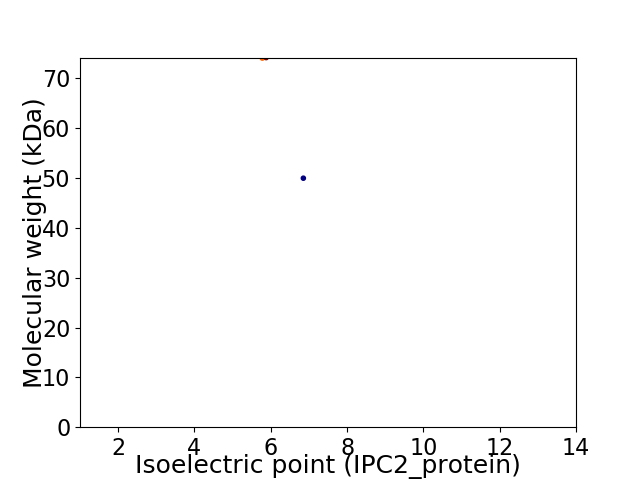
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEM1|A0A1L3KEM1_9VIRU RNA-directed RNA polymerase OS=Shahe sobemo-like virus 1 OX=1923454 PE=4 SV=1
MM1 pKa = 7.31GFIGIMASFFSLVVNVFFYY20 pKa = 9.87FSIFLAGALAVLMMEE35 pKa = 4.88GVQYY39 pKa = 10.28PFQNQIISAFVMYY52 pKa = 10.35IIVNGLLRR60 pKa = 11.84WRR62 pKa = 11.84RR63 pKa = 11.84SRR65 pKa = 11.84IPVIPAVLPQLDD77 pKa = 4.19PQDD80 pKa = 5.28LLDD83 pKa = 4.72LDD85 pKa = 4.42EE86 pKa = 5.48LLSEE90 pKa = 4.6ALPLVRR96 pKa = 11.84QRR98 pKa = 11.84HH99 pKa = 4.21SAIRR103 pKa = 11.84GLLTDD108 pKa = 3.55RR109 pKa = 11.84HH110 pKa = 6.26ALNKK114 pKa = 10.3LPEE117 pKa = 4.44GVFGIALGMPDD128 pKa = 4.34QNDD131 pKa = 3.21IEE133 pKa = 5.87DD134 pKa = 3.87NVFCGAVRR142 pKa = 11.84LGGFLVMNCHH152 pKa = 7.32AYY154 pKa = 9.14RR155 pKa = 11.84PKK157 pKa = 10.0MEE159 pKa = 4.08YY160 pKa = 8.36HH161 pKa = 5.35TTLVTKK167 pKa = 10.56RR168 pKa = 11.84GNFWMRR174 pKa = 11.84DD175 pKa = 3.44LVVTPDD181 pKa = 3.7PEE183 pKa = 4.28EE184 pKa = 4.23EE185 pKa = 4.15NVLEE189 pKa = 5.56DD190 pKa = 2.87MDD192 pKa = 5.1LFYY195 pKa = 11.21FRR197 pKa = 11.84LNPHH201 pKa = 6.42GLCSTKK207 pKa = 10.49AAEE210 pKa = 4.41TTVITQEE217 pKa = 4.24GVSARR222 pKa = 11.84IISLVNRR229 pKa = 11.84GVSSFGTVRR238 pKa = 11.84TFLEE242 pKa = 4.71GEE244 pKa = 4.24TTFGVVFYY252 pKa = 10.86SGKK255 pKa = 8.62TKK257 pKa = 10.21PGCSGFPYY265 pKa = 10.44LVGGKK270 pKa = 9.3VVGLHH275 pKa = 6.28CGDD278 pKa = 3.87YY279 pKa = 10.92NGTYY283 pKa = 10.74GDD285 pKa = 3.94GANVQLVLAKK295 pKa = 10.41LKK297 pKa = 10.6RR298 pKa = 11.84LSLGNRR304 pKa = 11.84HH305 pKa = 4.52SWFWVLDD312 pKa = 3.67DD313 pKa = 4.12RR314 pKa = 11.84HH315 pKa = 8.21AEE317 pKa = 3.93FTKK320 pKa = 9.99ISHH323 pKa = 6.35SGTPGEE329 pKa = 4.88LIVEE333 pKa = 4.15NSGRR337 pKa = 11.84YY338 pKa = 9.23YY339 pKa = 11.04YY340 pKa = 10.88VDD342 pKa = 3.22QEE344 pKa = 4.75DD345 pKa = 4.88YY346 pKa = 10.21LTEE349 pKa = 3.85ARR351 pKa = 11.84KK352 pKa = 10.16RR353 pKa = 11.84GVWAANHH360 pKa = 6.23KK361 pKa = 10.31GVFGRR366 pKa = 11.84FVGDD370 pKa = 3.37EE371 pKa = 4.09RR372 pKa = 11.84DD373 pKa = 3.58EE374 pKa = 3.98EE375 pKa = 5.14DD376 pKa = 3.64YY377 pKa = 11.69EE378 pKa = 4.13NDD380 pKa = 3.25RR381 pKa = 11.84RR382 pKa = 11.84SDD384 pKa = 3.51SDD386 pKa = 3.04HH387 pKa = 5.93GRR389 pKa = 11.84SRR391 pKa = 11.84SSARR395 pKa = 11.84SDD397 pKa = 3.87YY398 pKa = 11.46SDD400 pKa = 5.39DD401 pKa = 3.69DD402 pKa = 3.84HH403 pKa = 7.41FSNFEE408 pKa = 4.09DD409 pKa = 5.48DD410 pKa = 4.59EE411 pKa = 6.14FDD413 pKa = 4.05DD414 pKa = 4.34AHH416 pKa = 6.51SRR418 pKa = 11.84WVGRR422 pKa = 11.84QKK424 pKa = 10.85RR425 pKa = 11.84HH426 pKa = 5.13GKK428 pKa = 9.31SLSGFKK434 pKa = 10.06TGLNDD439 pKa = 4.31GNPDD443 pKa = 3.13SDD445 pKa = 3.82AVVSKK450 pKa = 10.78NVEE453 pKa = 4.0GPLPEE458 pKa = 4.87GPGYY462 pKa = 10.96AVTARR467 pKa = 11.84LRR469 pKa = 11.84FGADD473 pKa = 2.92PEE475 pKa = 3.95IRR477 pKa = 11.84MTSGEE482 pKa = 4.18IKK484 pKa = 10.52QEE486 pKa = 3.92AQEE489 pKa = 4.08IAACQKK495 pKa = 10.63KK496 pKa = 9.6IAEE499 pKa = 4.06LLARR503 pKa = 11.84VEE505 pKa = 4.14ALTTKK510 pKa = 10.45AQDD513 pKa = 3.29YY514 pKa = 9.9DD515 pKa = 3.62QLHH518 pKa = 5.36VQMEE522 pKa = 4.36KK523 pKa = 11.05DD524 pKa = 3.14RR525 pKa = 11.84VRR527 pKa = 11.84ALMEE531 pKa = 4.66AKK533 pKa = 10.24SQSDD537 pKa = 3.42QAAAQYY543 pKa = 9.11MVAISALKK551 pKa = 10.66SEE553 pKa = 4.26LAKK556 pKa = 10.65EE557 pKa = 3.82KK558 pKa = 10.86SEE560 pKa = 4.01RR561 pKa = 11.84LTTKK565 pKa = 10.77EE566 pKa = 4.46EE567 pKa = 3.88LHH569 pKa = 5.89QMHH572 pKa = 6.57LTVSEE577 pKa = 4.12LRR579 pKa = 11.84KK580 pKa = 10.01EE581 pKa = 4.05MRR583 pKa = 11.84SANARR588 pKa = 11.84LTEE591 pKa = 3.99AKK593 pKa = 10.52APVKK597 pKa = 10.22LKK599 pKa = 10.58PVVRR603 pKa = 11.84KK604 pKa = 6.53QTKK607 pKa = 8.46NEE609 pKa = 4.04KK610 pKa = 9.99KK611 pKa = 10.26VEE613 pKa = 4.04RR614 pKa = 11.84CLEE617 pKa = 4.37LGCMYY622 pKa = 10.33PKK624 pKa = 10.53EE625 pKa = 4.33ILLTMHH631 pKa = 7.01GPGLDD636 pKa = 3.45ALLQKK641 pKa = 10.89LEE643 pKa = 4.29ADD645 pKa = 4.38RR646 pKa = 11.84EE647 pKa = 4.23DD648 pKa = 3.91ASASRR653 pKa = 11.84QAGAIPRR660 pKa = 11.84NN661 pKa = 3.69
MM1 pKa = 7.31GFIGIMASFFSLVVNVFFYY20 pKa = 9.87FSIFLAGALAVLMMEE35 pKa = 4.88GVQYY39 pKa = 10.28PFQNQIISAFVMYY52 pKa = 10.35IIVNGLLRR60 pKa = 11.84WRR62 pKa = 11.84RR63 pKa = 11.84SRR65 pKa = 11.84IPVIPAVLPQLDD77 pKa = 4.19PQDD80 pKa = 5.28LLDD83 pKa = 4.72LDD85 pKa = 4.42EE86 pKa = 5.48LLSEE90 pKa = 4.6ALPLVRR96 pKa = 11.84QRR98 pKa = 11.84HH99 pKa = 4.21SAIRR103 pKa = 11.84GLLTDD108 pKa = 3.55RR109 pKa = 11.84HH110 pKa = 6.26ALNKK114 pKa = 10.3LPEE117 pKa = 4.44GVFGIALGMPDD128 pKa = 4.34QNDD131 pKa = 3.21IEE133 pKa = 5.87DD134 pKa = 3.87NVFCGAVRR142 pKa = 11.84LGGFLVMNCHH152 pKa = 7.32AYY154 pKa = 9.14RR155 pKa = 11.84PKK157 pKa = 10.0MEE159 pKa = 4.08YY160 pKa = 8.36HH161 pKa = 5.35TTLVTKK167 pKa = 10.56RR168 pKa = 11.84GNFWMRR174 pKa = 11.84DD175 pKa = 3.44LVVTPDD181 pKa = 3.7PEE183 pKa = 4.28EE184 pKa = 4.23EE185 pKa = 4.15NVLEE189 pKa = 5.56DD190 pKa = 2.87MDD192 pKa = 5.1LFYY195 pKa = 11.21FRR197 pKa = 11.84LNPHH201 pKa = 6.42GLCSTKK207 pKa = 10.49AAEE210 pKa = 4.41TTVITQEE217 pKa = 4.24GVSARR222 pKa = 11.84IISLVNRR229 pKa = 11.84GVSSFGTVRR238 pKa = 11.84TFLEE242 pKa = 4.71GEE244 pKa = 4.24TTFGVVFYY252 pKa = 10.86SGKK255 pKa = 8.62TKK257 pKa = 10.21PGCSGFPYY265 pKa = 10.44LVGGKK270 pKa = 9.3VVGLHH275 pKa = 6.28CGDD278 pKa = 3.87YY279 pKa = 10.92NGTYY283 pKa = 10.74GDD285 pKa = 3.94GANVQLVLAKK295 pKa = 10.41LKK297 pKa = 10.6RR298 pKa = 11.84LSLGNRR304 pKa = 11.84HH305 pKa = 4.52SWFWVLDD312 pKa = 3.67DD313 pKa = 4.12RR314 pKa = 11.84HH315 pKa = 8.21AEE317 pKa = 3.93FTKK320 pKa = 9.99ISHH323 pKa = 6.35SGTPGEE329 pKa = 4.88LIVEE333 pKa = 4.15NSGRR337 pKa = 11.84YY338 pKa = 9.23YY339 pKa = 11.04YY340 pKa = 10.88VDD342 pKa = 3.22QEE344 pKa = 4.75DD345 pKa = 4.88YY346 pKa = 10.21LTEE349 pKa = 3.85ARR351 pKa = 11.84KK352 pKa = 10.16RR353 pKa = 11.84GVWAANHH360 pKa = 6.23KK361 pKa = 10.31GVFGRR366 pKa = 11.84FVGDD370 pKa = 3.37EE371 pKa = 4.09RR372 pKa = 11.84DD373 pKa = 3.58EE374 pKa = 3.98EE375 pKa = 5.14DD376 pKa = 3.64YY377 pKa = 11.69EE378 pKa = 4.13NDD380 pKa = 3.25RR381 pKa = 11.84RR382 pKa = 11.84SDD384 pKa = 3.51SDD386 pKa = 3.04HH387 pKa = 5.93GRR389 pKa = 11.84SRR391 pKa = 11.84SSARR395 pKa = 11.84SDD397 pKa = 3.87YY398 pKa = 11.46SDD400 pKa = 5.39DD401 pKa = 3.69DD402 pKa = 3.84HH403 pKa = 7.41FSNFEE408 pKa = 4.09DD409 pKa = 5.48DD410 pKa = 4.59EE411 pKa = 6.14FDD413 pKa = 4.05DD414 pKa = 4.34AHH416 pKa = 6.51SRR418 pKa = 11.84WVGRR422 pKa = 11.84QKK424 pKa = 10.85RR425 pKa = 11.84HH426 pKa = 5.13GKK428 pKa = 9.31SLSGFKK434 pKa = 10.06TGLNDD439 pKa = 4.31GNPDD443 pKa = 3.13SDD445 pKa = 3.82AVVSKK450 pKa = 10.78NVEE453 pKa = 4.0GPLPEE458 pKa = 4.87GPGYY462 pKa = 10.96AVTARR467 pKa = 11.84LRR469 pKa = 11.84FGADD473 pKa = 2.92PEE475 pKa = 3.95IRR477 pKa = 11.84MTSGEE482 pKa = 4.18IKK484 pKa = 10.52QEE486 pKa = 3.92AQEE489 pKa = 4.08IAACQKK495 pKa = 10.63KK496 pKa = 9.6IAEE499 pKa = 4.06LLARR503 pKa = 11.84VEE505 pKa = 4.14ALTTKK510 pKa = 10.45AQDD513 pKa = 3.29YY514 pKa = 9.9DD515 pKa = 3.62QLHH518 pKa = 5.36VQMEE522 pKa = 4.36KK523 pKa = 11.05DD524 pKa = 3.14RR525 pKa = 11.84VRR527 pKa = 11.84ALMEE531 pKa = 4.66AKK533 pKa = 10.24SQSDD537 pKa = 3.42QAAAQYY543 pKa = 9.11MVAISALKK551 pKa = 10.66SEE553 pKa = 4.26LAKK556 pKa = 10.65EE557 pKa = 3.82KK558 pKa = 10.86SEE560 pKa = 4.01RR561 pKa = 11.84LTTKK565 pKa = 10.77EE566 pKa = 4.46EE567 pKa = 3.88LHH569 pKa = 5.89QMHH572 pKa = 6.57LTVSEE577 pKa = 4.12LRR579 pKa = 11.84KK580 pKa = 10.01EE581 pKa = 4.05MRR583 pKa = 11.84SANARR588 pKa = 11.84LTEE591 pKa = 3.99AKK593 pKa = 10.52APVKK597 pKa = 10.22LKK599 pKa = 10.58PVVRR603 pKa = 11.84KK604 pKa = 6.53QTKK607 pKa = 8.46NEE609 pKa = 4.04KK610 pKa = 9.99KK611 pKa = 10.26VEE613 pKa = 4.04RR614 pKa = 11.84CLEE617 pKa = 4.37LGCMYY622 pKa = 10.33PKK624 pKa = 10.53EE625 pKa = 4.33ILLTMHH631 pKa = 7.01GPGLDD636 pKa = 3.45ALLQKK641 pKa = 10.89LEE643 pKa = 4.29ADD645 pKa = 4.38RR646 pKa = 11.84EE647 pKa = 4.23DD648 pKa = 3.91ASASRR653 pKa = 11.84QAGAIPRR660 pKa = 11.84NN661 pKa = 3.69
Molecular weight: 74.07 kDa
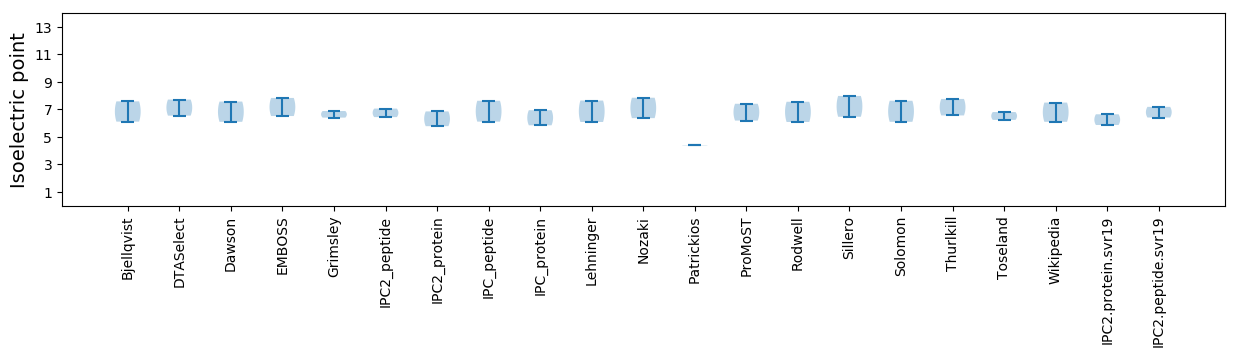
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEM1|A0A1L3KEM1_9VIRU RNA-directed RNA polymerase OS=Shahe sobemo-like virus 1 OX=1923454 PE=4 SV=1
MM1 pKa = 7.53FDD3 pKa = 3.01WPNRR7 pKa = 11.84NKK9 pKa = 10.73AAVDD13 pKa = 3.38EE14 pKa = 4.7SVRR17 pKa = 11.84QHH19 pKa = 5.43VKK21 pKa = 10.39HH22 pKa = 6.83RR23 pKa = 11.84DD24 pKa = 3.37DD25 pKa = 4.07VADD28 pKa = 3.49KK29 pKa = 9.98TVAPTRR35 pKa = 11.84EE36 pKa = 4.27EE37 pKa = 3.82YY38 pKa = 10.89SEE40 pKa = 4.01ILEE43 pKa = 4.26MTLKK47 pKa = 10.85AFGRR51 pKa = 11.84GWSLFNPDD59 pKa = 4.3EE60 pKa = 4.46ISVIRR65 pKa = 11.84IRR67 pKa = 11.84EE68 pKa = 4.19IIEE71 pKa = 3.94HH72 pKa = 5.58QVDD75 pKa = 3.92KK76 pKa = 10.94NSSPGYY82 pKa = 9.2PYY84 pKa = 10.75CLKK87 pKa = 10.21TKK89 pKa = 7.93TNKK92 pKa = 10.3DD93 pKa = 3.25FFGFDD98 pKa = 3.01ALGRR102 pKa = 11.84VPDD105 pKa = 4.28DD106 pKa = 3.79RR107 pKa = 11.84LAMVCGLVADD117 pKa = 6.05RR118 pKa = 11.84IDD120 pKa = 3.79EE121 pKa = 4.37LATRR125 pKa = 11.84PASDD129 pKa = 5.1PIRR132 pKa = 11.84VFIKK136 pKa = 10.46EE137 pKa = 4.03EE138 pKa = 3.54PASINKK144 pKa = 9.77RR145 pKa = 11.84STGALRR151 pKa = 11.84LISSVSVVDD160 pKa = 4.45TIVHH164 pKa = 6.29RR165 pKa = 11.84LFFGGVFDD173 pKa = 5.62KK174 pKa = 11.12LVKK177 pKa = 10.21HH178 pKa = 6.47WGATPNMAGWSLGGGKK194 pKa = 8.25WRR196 pKa = 11.84FFKK199 pKa = 10.75KK200 pKa = 9.87HH201 pKa = 5.36FKK203 pKa = 10.62GKK205 pKa = 10.77AFMADD210 pKa = 2.92KK211 pKa = 10.84KK212 pKa = 10.7KK213 pKa = 9.9WDD215 pKa = 3.42WSMQPWLANLTRR227 pKa = 11.84DD228 pKa = 3.25VLLNLGRR235 pKa = 11.84FEE237 pKa = 3.85GRR239 pKa = 11.84EE240 pKa = 3.6RR241 pKa = 11.84VIADD245 pKa = 3.2NALRR249 pKa = 11.84ALYY252 pKa = 10.42GMKK255 pKa = 10.28FEE257 pKa = 4.62GKK259 pKa = 9.76NYY261 pKa = 7.36PTTLVSACTRR271 pKa = 11.84CGACSFYY278 pKa = 10.3EE279 pKa = 3.86QRR281 pKa = 11.84TSGLQKK287 pKa = 10.63SGWLGTIITNGMQQYY302 pKa = 11.29ALAALAKK309 pKa = 9.43RR310 pKa = 11.84RR311 pKa = 11.84LGRR314 pKa = 11.84SMGPIFTMGDD324 pKa = 3.28DD325 pKa = 4.14TIEE328 pKa = 4.17EE329 pKa = 4.28DD330 pKa = 3.75VDD332 pKa = 4.42DD333 pKa = 4.86VEE335 pKa = 5.06APIYY339 pKa = 10.54VSEE342 pKa = 4.44LEE344 pKa = 4.12KK345 pKa = 10.95GGCIVGEE352 pKa = 4.18YY353 pKa = 10.45KK354 pKa = 9.69STEE357 pKa = 4.16DD358 pKa = 3.34YY359 pKa = 11.51DD360 pKa = 3.92FAGFTFPNGGGRR372 pKa = 11.84PVPNYY377 pKa = 10.01GDD379 pKa = 3.09KK380 pKa = 10.95HH381 pKa = 6.84AFTLAYY387 pKa = 10.23ANDD390 pKa = 3.97KK391 pKa = 10.82EE392 pKa = 5.17GVFADD397 pKa = 3.8EE398 pKa = 4.24QSRR401 pKa = 11.84VASYY405 pKa = 8.93ITMYY409 pKa = 11.15YY410 pKa = 9.72NDD412 pKa = 3.64KK413 pKa = 10.99ASYY416 pKa = 10.36RR417 pKa = 11.84KK418 pKa = 9.46LLQYY422 pKa = 11.12AAVLEE427 pKa = 4.3VDD429 pKa = 3.68QVPSDD434 pKa = 3.8FEE436 pKa = 4.29LMRR439 pKa = 11.84WYY441 pKa = 10.68GG442 pKa = 3.27
MM1 pKa = 7.53FDD3 pKa = 3.01WPNRR7 pKa = 11.84NKK9 pKa = 10.73AAVDD13 pKa = 3.38EE14 pKa = 4.7SVRR17 pKa = 11.84QHH19 pKa = 5.43VKK21 pKa = 10.39HH22 pKa = 6.83RR23 pKa = 11.84DD24 pKa = 3.37DD25 pKa = 4.07VADD28 pKa = 3.49KK29 pKa = 9.98TVAPTRR35 pKa = 11.84EE36 pKa = 4.27EE37 pKa = 3.82YY38 pKa = 10.89SEE40 pKa = 4.01ILEE43 pKa = 4.26MTLKK47 pKa = 10.85AFGRR51 pKa = 11.84GWSLFNPDD59 pKa = 4.3EE60 pKa = 4.46ISVIRR65 pKa = 11.84IRR67 pKa = 11.84EE68 pKa = 4.19IIEE71 pKa = 3.94HH72 pKa = 5.58QVDD75 pKa = 3.92KK76 pKa = 10.94NSSPGYY82 pKa = 9.2PYY84 pKa = 10.75CLKK87 pKa = 10.21TKK89 pKa = 7.93TNKK92 pKa = 10.3DD93 pKa = 3.25FFGFDD98 pKa = 3.01ALGRR102 pKa = 11.84VPDD105 pKa = 4.28DD106 pKa = 3.79RR107 pKa = 11.84LAMVCGLVADD117 pKa = 6.05RR118 pKa = 11.84IDD120 pKa = 3.79EE121 pKa = 4.37LATRR125 pKa = 11.84PASDD129 pKa = 5.1PIRR132 pKa = 11.84VFIKK136 pKa = 10.46EE137 pKa = 4.03EE138 pKa = 3.54PASINKK144 pKa = 9.77RR145 pKa = 11.84STGALRR151 pKa = 11.84LISSVSVVDD160 pKa = 4.45TIVHH164 pKa = 6.29RR165 pKa = 11.84LFFGGVFDD173 pKa = 5.62KK174 pKa = 11.12LVKK177 pKa = 10.21HH178 pKa = 6.47WGATPNMAGWSLGGGKK194 pKa = 8.25WRR196 pKa = 11.84FFKK199 pKa = 10.75KK200 pKa = 9.87HH201 pKa = 5.36FKK203 pKa = 10.62GKK205 pKa = 10.77AFMADD210 pKa = 2.92KK211 pKa = 10.84KK212 pKa = 10.7KK213 pKa = 9.9WDD215 pKa = 3.42WSMQPWLANLTRR227 pKa = 11.84DD228 pKa = 3.25VLLNLGRR235 pKa = 11.84FEE237 pKa = 3.85GRR239 pKa = 11.84EE240 pKa = 3.6RR241 pKa = 11.84VIADD245 pKa = 3.2NALRR249 pKa = 11.84ALYY252 pKa = 10.42GMKK255 pKa = 10.28FEE257 pKa = 4.62GKK259 pKa = 9.76NYY261 pKa = 7.36PTTLVSACTRR271 pKa = 11.84CGACSFYY278 pKa = 10.3EE279 pKa = 3.86QRR281 pKa = 11.84TSGLQKK287 pKa = 10.63SGWLGTIITNGMQQYY302 pKa = 11.29ALAALAKK309 pKa = 9.43RR310 pKa = 11.84RR311 pKa = 11.84LGRR314 pKa = 11.84SMGPIFTMGDD324 pKa = 3.28DD325 pKa = 4.14TIEE328 pKa = 4.17EE329 pKa = 4.28DD330 pKa = 3.75VDD332 pKa = 4.42DD333 pKa = 4.86VEE335 pKa = 5.06APIYY339 pKa = 10.54VSEE342 pKa = 4.44LEE344 pKa = 4.12KK345 pKa = 10.95GGCIVGEE352 pKa = 4.18YY353 pKa = 10.45KK354 pKa = 9.69STEE357 pKa = 4.16DD358 pKa = 3.34YY359 pKa = 11.51DD360 pKa = 3.92FAGFTFPNGGGRR372 pKa = 11.84PVPNYY377 pKa = 10.01GDD379 pKa = 3.09KK380 pKa = 10.95HH381 pKa = 6.84AFTLAYY387 pKa = 10.23ANDD390 pKa = 3.97KK391 pKa = 10.82EE392 pKa = 5.17GVFADD397 pKa = 3.8EE398 pKa = 4.24QSRR401 pKa = 11.84VASYY405 pKa = 8.93ITMYY409 pKa = 11.15YY410 pKa = 9.72NDD412 pKa = 3.64KK413 pKa = 10.99ASYY416 pKa = 10.36RR417 pKa = 11.84KK418 pKa = 9.46LLQYY422 pKa = 11.12AAVLEE427 pKa = 4.3VDD429 pKa = 3.68QVPSDD434 pKa = 3.8FEE436 pKa = 4.29LMRR439 pKa = 11.84WYY441 pKa = 10.68GG442 pKa = 3.27
Molecular weight: 49.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1103 |
442 |
661 |
551.5 |
62.02 |
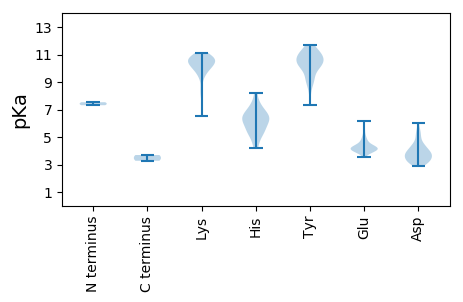
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.978 ± 0.103 | 1.269 ± 0.055 |
6.89 ± 0.356 | 6.8 ± 0.567 |
4.805 ± 0.387 | 8.069 ± 0.187 |
2.267 ± 0.422 | 3.989 ± 0.331 |
5.984 ± 0.497 | 8.794 ± 0.962 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.72 ± 0.003 | 3.626 ± 0.004 |
3.898 ± 0.108 | 3.083 ± 0.507 |
6.981 ± 0.12 | 6.074 ± 0.259 |
4.624 ± 0.219 | 7.253 ± 0.288 |
1.451 ± 0.502 | 3.445 ± 0.388 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
