
Streptococcus phage Javan497
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.7
Get precalculated fractions of proteins
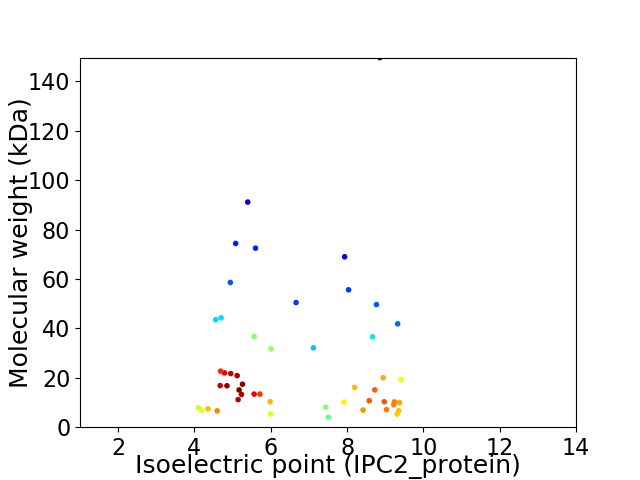
Virtual 2D-PAGE plot for 50 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4D6AJA8|A0A4D6AJA8_9CAUD Transcriptional activator OS=Streptococcus phage Javan497 OX=2548211 GN=Javan497_0022 PE=4 SV=1
MM1 pKa = 7.64NYY3 pKa = 10.44LEE5 pKa = 4.33YY6 pKa = 11.13ALAYY10 pKa = 10.27LEE12 pKa = 4.77RR13 pKa = 11.84EE14 pKa = 4.2LEE16 pKa = 4.54IIDD19 pKa = 4.43DD20 pKa = 3.92EE21 pKa = 4.88VIEE24 pKa = 4.2VEE26 pKa = 4.76LPGGDD31 pKa = 3.23WEE33 pKa = 4.69FVPNPYY39 pKa = 10.03YY40 pKa = 10.52EE41 pKa = 4.88EE42 pKa = 4.6GLHH45 pKa = 6.64DD46 pKa = 3.79SPYY49 pKa = 10.48YY50 pKa = 10.02RR51 pKa = 11.84SQVAKK56 pKa = 10.55DD57 pKa = 3.09ILDD60 pKa = 3.46IKK62 pKa = 10.8GLLGRR67 pKa = 4.72
MM1 pKa = 7.64NYY3 pKa = 10.44LEE5 pKa = 4.33YY6 pKa = 11.13ALAYY10 pKa = 10.27LEE12 pKa = 4.77RR13 pKa = 11.84EE14 pKa = 4.2LEE16 pKa = 4.54IIDD19 pKa = 4.43DD20 pKa = 3.92EE21 pKa = 4.88VIEE24 pKa = 4.2VEE26 pKa = 4.76LPGGDD31 pKa = 3.23WEE33 pKa = 4.69FVPNPYY39 pKa = 10.03YY40 pKa = 10.52EE41 pKa = 4.88EE42 pKa = 4.6GLHH45 pKa = 6.64DD46 pKa = 3.79SPYY49 pKa = 10.48YY50 pKa = 10.02RR51 pKa = 11.84SQVAKK56 pKa = 10.55DD57 pKa = 3.09ILDD60 pKa = 3.46IKK62 pKa = 10.8GLLGRR67 pKa = 4.72
Molecular weight: 7.87 kDa
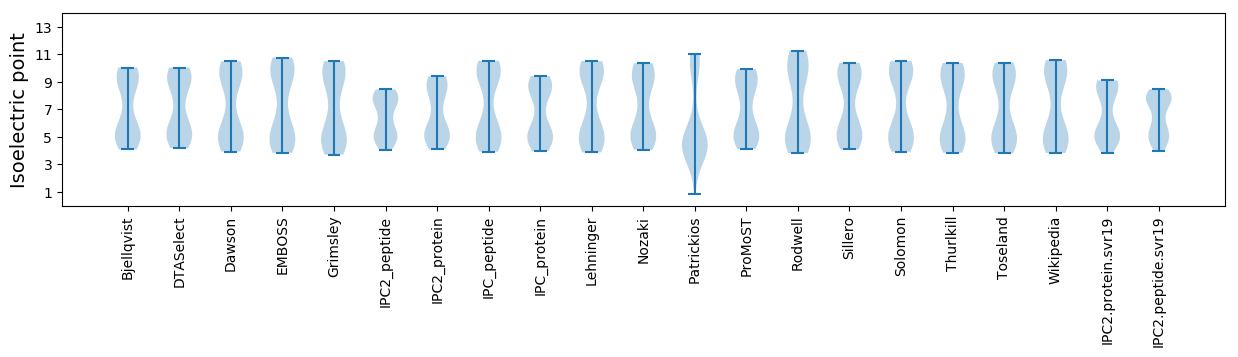
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D6AHH3|A0A4D6AHH3_9CAUD Uncharacterized protein OS=Streptococcus phage Javan497 OX=2548211 GN=Javan497_0011 PE=4 SV=1
MM1 pKa = 7.32IEE3 pKa = 5.0KK4 pKa = 8.7YY5 pKa = 8.55TKK7 pKa = 10.41KK8 pKa = 10.57DD9 pKa = 2.72GTTAYY14 pKa = 10.65RR15 pKa = 11.84LRR17 pKa = 11.84AYY19 pKa = 10.71LGVDD23 pKa = 3.48PMTGKK28 pKa = 9.66QVRR31 pKa = 11.84TTRR34 pKa = 11.84QGFKK38 pKa = 9.38TEE40 pKa = 4.06RR41 pKa = 11.84EE42 pKa = 4.18AKK44 pKa = 8.09RR45 pKa = 11.84TEE47 pKa = 4.01VKK49 pKa = 10.68LVDD52 pKa = 3.62DD53 pKa = 5.01FQRR56 pKa = 11.84QGAWKK61 pKa = 10.31NNDD64 pKa = 2.62KK65 pKa = 8.55TTFDD69 pKa = 3.5DD70 pKa = 5.1AARR73 pKa = 11.84LWFEE77 pKa = 3.98QYY79 pKa = 11.18KK80 pKa = 8.56NTVKK84 pKa = 10.76PSTFLVNQNYY94 pKa = 9.38YY95 pKa = 10.64KK96 pKa = 10.0STIKK100 pKa = 10.34PHH102 pKa = 6.22IGKK105 pKa = 8.65SQISKK110 pKa = 7.62ITVMACQNLINHH122 pKa = 7.46LAQYY126 pKa = 8.0SACASYY132 pKa = 10.74ISVINRR138 pKa = 11.84IFKK141 pKa = 10.19FAVHH145 pKa = 6.62LGIIEE150 pKa = 4.38NNPMDD155 pKa = 3.51RR156 pKa = 11.84TIRR159 pKa = 11.84ARR161 pKa = 11.84CTYY164 pKa = 9.73AHH166 pKa = 6.98KK167 pKa = 11.03EE168 pKa = 3.59KK169 pKa = 10.24MFYY172 pKa = 10.59SKK174 pKa = 11.32EE175 pKa = 3.87EE176 pKa = 3.83LLIFLDD182 pKa = 3.65IVKK185 pKa = 10.25KK186 pKa = 10.63EE187 pKa = 4.05EE188 pKa = 4.17SLEE191 pKa = 3.75MLLIYY196 pKa = 10.48RR197 pKa = 11.84LLAYY201 pKa = 10.06GGLRR205 pKa = 11.84IGEE208 pKa = 4.13AVALYY213 pKa = 8.48DD214 pKa = 4.0TDD216 pKa = 5.82FDD218 pKa = 4.2FTNNTISITKK228 pKa = 9.07TLAQTKK234 pKa = 9.56AGKK237 pKa = 10.07VIQNPKK243 pKa = 7.57TKK245 pKa = 10.04KK246 pKa = 9.1SSRR249 pKa = 11.84TISMDD254 pKa = 3.19QEE256 pKa = 4.1TMRR259 pKa = 11.84LAKK262 pKa = 10.13EE263 pKa = 3.84YY264 pKa = 9.12MRR266 pKa = 11.84QSVRR270 pKa = 11.84PLHH273 pKa = 6.09GSFRR277 pKa = 11.84VFNLLTSTVRR287 pKa = 11.84RR288 pKa = 11.84RR289 pKa = 11.84LKK291 pKa = 9.44RR292 pKa = 11.84TVEE295 pKa = 3.72RR296 pKa = 11.84HH297 pKa = 4.98HH298 pKa = 6.73LKK300 pKa = 10.45KK301 pKa = 10.44INPHH305 pKa = 5.44GFRR308 pKa = 11.84HH309 pKa = 4.99THH311 pKa = 6.0ASLLFEE317 pKa = 5.22AGVPAKK323 pKa = 10.26LAQEE327 pKa = 4.26RR328 pKa = 11.84LGHH331 pKa = 5.98AKK333 pKa = 10.11ISITMDD339 pKa = 3.7LYY341 pKa = 10.51THH343 pKa = 7.22LSKK346 pKa = 11.23NQTDD350 pKa = 3.66NVAEE354 pKa = 4.12KK355 pKa = 10.42LADD358 pKa = 5.17FIAII362 pKa = 4.09
MM1 pKa = 7.32IEE3 pKa = 5.0KK4 pKa = 8.7YY5 pKa = 8.55TKK7 pKa = 10.41KK8 pKa = 10.57DD9 pKa = 2.72GTTAYY14 pKa = 10.65RR15 pKa = 11.84LRR17 pKa = 11.84AYY19 pKa = 10.71LGVDD23 pKa = 3.48PMTGKK28 pKa = 9.66QVRR31 pKa = 11.84TTRR34 pKa = 11.84QGFKK38 pKa = 9.38TEE40 pKa = 4.06RR41 pKa = 11.84EE42 pKa = 4.18AKK44 pKa = 8.09RR45 pKa = 11.84TEE47 pKa = 4.01VKK49 pKa = 10.68LVDD52 pKa = 3.62DD53 pKa = 5.01FQRR56 pKa = 11.84QGAWKK61 pKa = 10.31NNDD64 pKa = 2.62KK65 pKa = 8.55TTFDD69 pKa = 3.5DD70 pKa = 5.1AARR73 pKa = 11.84LWFEE77 pKa = 3.98QYY79 pKa = 11.18KK80 pKa = 8.56NTVKK84 pKa = 10.76PSTFLVNQNYY94 pKa = 9.38YY95 pKa = 10.64KK96 pKa = 10.0STIKK100 pKa = 10.34PHH102 pKa = 6.22IGKK105 pKa = 8.65SQISKK110 pKa = 7.62ITVMACQNLINHH122 pKa = 7.46LAQYY126 pKa = 8.0SACASYY132 pKa = 10.74ISVINRR138 pKa = 11.84IFKK141 pKa = 10.19FAVHH145 pKa = 6.62LGIIEE150 pKa = 4.38NNPMDD155 pKa = 3.51RR156 pKa = 11.84TIRR159 pKa = 11.84ARR161 pKa = 11.84CTYY164 pKa = 9.73AHH166 pKa = 6.98KK167 pKa = 11.03EE168 pKa = 3.59KK169 pKa = 10.24MFYY172 pKa = 10.59SKK174 pKa = 11.32EE175 pKa = 3.87EE176 pKa = 3.83LLIFLDD182 pKa = 3.65IVKK185 pKa = 10.25KK186 pKa = 10.63EE187 pKa = 4.05EE188 pKa = 4.17SLEE191 pKa = 3.75MLLIYY196 pKa = 10.48RR197 pKa = 11.84LLAYY201 pKa = 10.06GGLRR205 pKa = 11.84IGEE208 pKa = 4.13AVALYY213 pKa = 8.48DD214 pKa = 4.0TDD216 pKa = 5.82FDD218 pKa = 4.2FTNNTISITKK228 pKa = 9.07TLAQTKK234 pKa = 9.56AGKK237 pKa = 10.07VIQNPKK243 pKa = 7.57TKK245 pKa = 10.04KK246 pKa = 9.1SSRR249 pKa = 11.84TISMDD254 pKa = 3.19QEE256 pKa = 4.1TMRR259 pKa = 11.84LAKK262 pKa = 10.13EE263 pKa = 3.84YY264 pKa = 9.12MRR266 pKa = 11.84QSVRR270 pKa = 11.84PLHH273 pKa = 6.09GSFRR277 pKa = 11.84VFNLLTSTVRR287 pKa = 11.84RR288 pKa = 11.84RR289 pKa = 11.84LKK291 pKa = 9.44RR292 pKa = 11.84TVEE295 pKa = 3.72RR296 pKa = 11.84HH297 pKa = 4.98HH298 pKa = 6.73LKK300 pKa = 10.45KK301 pKa = 10.44INPHH305 pKa = 5.44GFRR308 pKa = 11.84HH309 pKa = 4.99THH311 pKa = 6.0ASLLFEE317 pKa = 5.22AGVPAKK323 pKa = 10.26LAQEE327 pKa = 4.26RR328 pKa = 11.84LGHH331 pKa = 5.98AKK333 pKa = 10.11ISITMDD339 pKa = 3.7LYY341 pKa = 10.51THH343 pKa = 7.22LSKK346 pKa = 11.23NQTDD350 pKa = 3.66NVAEE354 pKa = 4.12KK355 pKa = 10.42LADD358 pKa = 5.17FIAII362 pKa = 4.09
Molecular weight: 41.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12102 |
38 |
1460 |
242.0 |
27.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.147 ± 0.646 | 0.463 ± 0.106 |
6.148 ± 0.488 | 7.238 ± 0.617 |
3.685 ± 0.175 | 6.453 ± 0.434 |
1.231 ± 0.172 | 6.949 ± 0.377 |
8.651 ± 0.484 | 8.792 ± 0.269 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.355 ± 0.143 | 5.197 ± 0.255 |
2.661 ± 0.195 | 3.867 ± 0.277 |
4.338 ± 0.387 | 6.561 ± 0.721 |
6.313 ± 0.29 | 6.148 ± 0.194 |
0.942 ± 0.144 | 3.859 ± 0.427 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
