
Sporomusa sphaeroides DSM 2875
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Negativicutes; Selenomonadales; Sporomusaceae; Sporomusa; Sporomusa sphaeroides
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
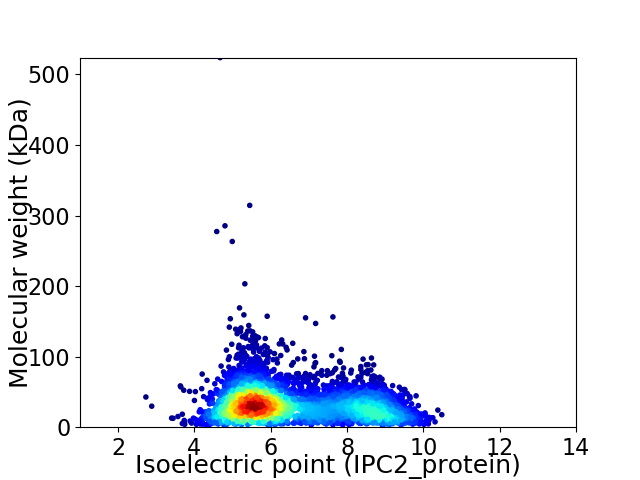
Virtual 2D-PAGE plot for 4565 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1U7MHL0|A0A1U7MHL0_9FIRM Germination-specific N-acetylmuramoyl-L-alanine amidase OS=Sporomusa sphaeroides DSM 2875 OX=1337886 GN=cwlD PE=4 SV=1
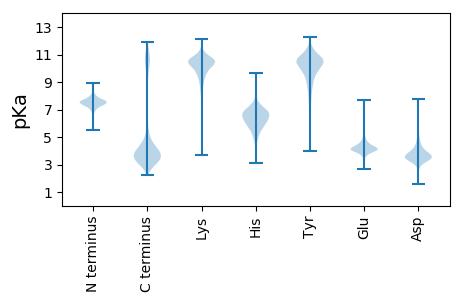
MM1 pKa = 7.57GSTFGGYY8 pKa = 9.91SIAYY12 pKa = 9.08SGMYY16 pKa = 10.24ANQSALTVTSQNLSNVNTSGYY37 pKa = 10.41SRR39 pKa = 11.84QQINSAEE46 pKa = 4.17KK47 pKa = 9.62IVPISSQTSIGAGVAVVEE65 pKa = 4.48VCRR68 pKa = 11.84ARR70 pKa = 11.84NQLLDD75 pKa = 3.56STYY78 pKa = 10.57HH79 pKa = 6.92DD80 pKa = 4.33KK81 pKa = 11.17NASASYY87 pKa = 9.24WSTKK91 pKa = 10.66SGMLTYY97 pKa = 10.39AQEE100 pKa = 4.2ILDD103 pKa = 4.4EE104 pKa = 4.98FSATDD109 pKa = 3.8GSGSDD114 pKa = 4.09GLQQTIQNFFDD125 pKa = 3.39SWEE128 pKa = 4.08EE129 pKa = 3.75LAGDD133 pKa = 4.65AGNIANRR140 pKa = 11.84QAVVEE145 pKa = 4.13YY146 pKa = 10.49AATLLDD152 pKa = 3.81TLSSIDD158 pKa = 3.62EE159 pKa = 4.04QLAALQQDD167 pKa = 4.12ACNSAADD174 pKa = 3.62IVDD177 pKa = 5.22DD178 pKa = 4.39INSIANQVAALNLQIVRR195 pKa = 11.84AEE197 pKa = 4.0AADD200 pKa = 3.97AEE202 pKa = 4.42ASDD205 pKa = 5.64LRR207 pKa = 11.84DD208 pKa = 3.37QRR210 pKa = 11.84DD211 pKa = 3.47YY212 pKa = 11.88LLDD215 pKa = 3.59QLSDD219 pKa = 3.57YY220 pKa = 11.7VNFSTQEE227 pKa = 3.75QANGSVTVFIGGVALVSLNKK247 pKa = 10.27SYY249 pKa = 10.79EE250 pKa = 3.85LSLAGDD256 pKa = 4.61GSTADD261 pKa = 3.63PLVVRR266 pKa = 11.84WADD269 pKa = 3.04TGTRR273 pKa = 11.84ASITNGTLKK282 pKa = 10.81ACLEE286 pKa = 4.61DD287 pKa = 4.61ADD289 pKa = 3.98QTGIGAIDD297 pKa = 3.93PEE299 pKa = 5.45LILDD303 pKa = 4.05TDD305 pKa = 3.45QDD307 pKa = 4.11GKK309 pKa = 11.38VYY311 pKa = 9.97TYY313 pKa = 8.78TASATSSLSNLRR325 pKa = 11.84QGLNTLITTLAVKK338 pKa = 10.53LNSLHH343 pKa = 6.61SAGTGLDD350 pKa = 3.47GSTGTDD356 pKa = 3.43FFVAIDD362 pKa = 3.72SSQPLSSTNIQVNPAIVSDD381 pKa = 4.23LNKK384 pKa = 10.16LAAGASGAAGDD395 pKa = 4.0NTVAAAICDD404 pKa = 3.86LSSAGIYY411 pKa = 9.72TFDD414 pKa = 4.8GLSQDD419 pKa = 2.89GTAFYY424 pKa = 10.82QSLISWLSTAGSDD437 pKa = 4.18ASDD440 pKa = 4.27YY441 pKa = 11.48YY442 pKa = 10.71DD443 pKa = 3.6TQSALATQVDD453 pKa = 3.95NQRR456 pKa = 11.84QSISTVSLDD465 pKa = 3.52EE466 pKa = 4.53EE467 pKa = 4.47MSNMITFQNAYY478 pKa = 9.12AASARR483 pKa = 11.84VLSVMDD489 pKa = 4.45GLLADD494 pKa = 5.86LIAEE498 pKa = 4.74LGG500 pKa = 3.57
MM1 pKa = 7.57GSTFGGYY8 pKa = 9.91SIAYY12 pKa = 9.08SGMYY16 pKa = 10.24ANQSALTVTSQNLSNVNTSGYY37 pKa = 10.41SRR39 pKa = 11.84QQINSAEE46 pKa = 4.17KK47 pKa = 9.62IVPISSQTSIGAGVAVVEE65 pKa = 4.48VCRR68 pKa = 11.84ARR70 pKa = 11.84NQLLDD75 pKa = 3.56STYY78 pKa = 10.57HH79 pKa = 6.92DD80 pKa = 4.33KK81 pKa = 11.17NASASYY87 pKa = 9.24WSTKK91 pKa = 10.66SGMLTYY97 pKa = 10.39AQEE100 pKa = 4.2ILDD103 pKa = 4.4EE104 pKa = 4.98FSATDD109 pKa = 3.8GSGSDD114 pKa = 4.09GLQQTIQNFFDD125 pKa = 3.39SWEE128 pKa = 4.08EE129 pKa = 3.75LAGDD133 pKa = 4.65AGNIANRR140 pKa = 11.84QAVVEE145 pKa = 4.13YY146 pKa = 10.49AATLLDD152 pKa = 3.81TLSSIDD158 pKa = 3.62EE159 pKa = 4.04QLAALQQDD167 pKa = 4.12ACNSAADD174 pKa = 3.62IVDD177 pKa = 5.22DD178 pKa = 4.39INSIANQVAALNLQIVRR195 pKa = 11.84AEE197 pKa = 4.0AADD200 pKa = 3.97AEE202 pKa = 4.42ASDD205 pKa = 5.64LRR207 pKa = 11.84DD208 pKa = 3.37QRR210 pKa = 11.84DD211 pKa = 3.47YY212 pKa = 11.88LLDD215 pKa = 3.59QLSDD219 pKa = 3.57YY220 pKa = 11.7VNFSTQEE227 pKa = 3.75QANGSVTVFIGGVALVSLNKK247 pKa = 10.27SYY249 pKa = 10.79EE250 pKa = 3.85LSLAGDD256 pKa = 4.61GSTADD261 pKa = 3.63PLVVRR266 pKa = 11.84WADD269 pKa = 3.04TGTRR273 pKa = 11.84ASITNGTLKK282 pKa = 10.81ACLEE286 pKa = 4.61DD287 pKa = 4.61ADD289 pKa = 3.98QTGIGAIDD297 pKa = 3.93PEE299 pKa = 5.45LILDD303 pKa = 4.05TDD305 pKa = 3.45QDD307 pKa = 4.11GKK309 pKa = 11.38VYY311 pKa = 9.97TYY313 pKa = 8.78TASATSSLSNLRR325 pKa = 11.84QGLNTLITTLAVKK338 pKa = 10.53LNSLHH343 pKa = 6.61SAGTGLDD350 pKa = 3.47GSTGTDD356 pKa = 3.43FFVAIDD362 pKa = 3.72SSQPLSSTNIQVNPAIVSDD381 pKa = 4.23LNKK384 pKa = 10.16LAAGASGAAGDD395 pKa = 4.0NTVAAAICDD404 pKa = 3.86LSSAGIYY411 pKa = 9.72TFDD414 pKa = 4.8GLSQDD419 pKa = 2.89GTAFYY424 pKa = 10.82QSLISWLSTAGSDD437 pKa = 4.18ASDD440 pKa = 4.27YY441 pKa = 11.48YY442 pKa = 10.71DD443 pKa = 3.6TQSALATQVDD453 pKa = 3.95NQRR456 pKa = 11.84QSISTVSLDD465 pKa = 3.52EE466 pKa = 4.53EE467 pKa = 4.47MSNMITFQNAYY478 pKa = 9.12AASARR483 pKa = 11.84VLSVMDD489 pKa = 4.45GLLADD494 pKa = 5.86LIAEE498 pKa = 4.74LGG500 pKa = 3.57
Molecular weight: 52.39 kDa
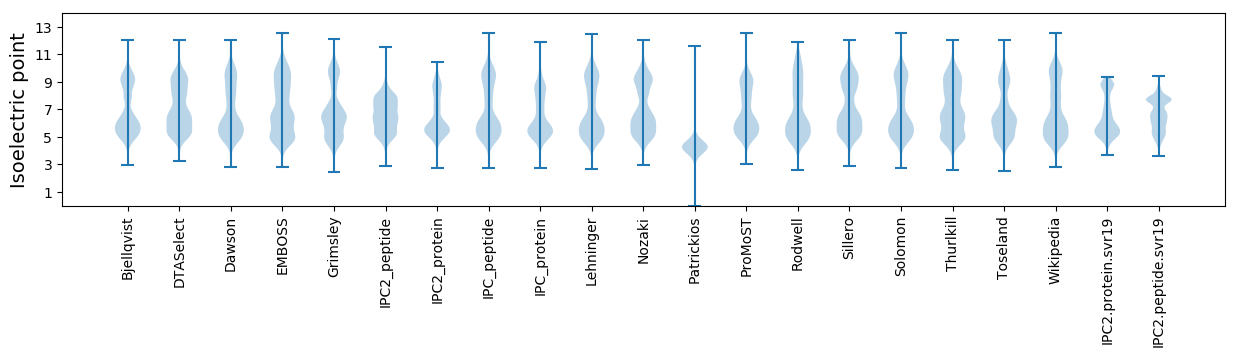
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1U7MJN6|A0A1U7MJN6_9FIRM Demethylrebeccamycin-D-glucose O-methyltransferase OS=Sporomusa sphaeroides DSM 2875 OX=1337886 GN=rebM_2 PE=4 SV=1
MM1 pKa = 7.62RR2 pKa = 11.84FEE4 pKa = 4.33MLLIIAGMAAVTFFTRR20 pKa = 11.84FGALALLKK28 pKa = 9.87KK29 pKa = 9.19TGLPCWFEE37 pKa = 3.2RR38 pKa = 11.84WLRR41 pKa = 11.84HH42 pKa = 5.27VPTAVLTALIVPALLLPQGRR62 pKa = 11.84VDD64 pKa = 4.54LSLSNHH70 pKa = 5.76YY71 pKa = 10.53LLAGILAAIVAYY83 pKa = 7.67TCRR86 pKa = 11.84NAMLTMGLGLTAMLSLRR103 pKa = 11.84WLGII107 pKa = 3.56
MM1 pKa = 7.62RR2 pKa = 11.84FEE4 pKa = 4.33MLLIIAGMAAVTFFTRR20 pKa = 11.84FGALALLKK28 pKa = 9.87KK29 pKa = 9.19TGLPCWFEE37 pKa = 3.2RR38 pKa = 11.84WLRR41 pKa = 11.84HH42 pKa = 5.27VPTAVLTALIVPALLLPQGRR62 pKa = 11.84VDD64 pKa = 4.54LSLSNHH70 pKa = 5.76YY71 pKa = 10.53LLAGILAAIVAYY83 pKa = 7.67TCRR86 pKa = 11.84NAMLTMGLGLTAMLSLRR103 pKa = 11.84WLGII107 pKa = 3.56
Molecular weight: 11.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1425425 |
29 |
5143 |
312.3 |
34.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.758 ± 0.045 | 1.233 ± 0.014 |
4.851 ± 0.026 | 6.173 ± 0.034 |
3.766 ± 0.027 | 7.667 ± 0.043 |
1.828 ± 0.015 | 7.19 ± 0.036 |
5.581 ± 0.031 | 9.963 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.679 ± 0.021 | 3.938 ± 0.036 |
3.982 ± 0.024 | 3.929 ± 0.027 |
4.742 ± 0.026 | 5.407 ± 0.027 |
5.622 ± 0.033 | 7.451 ± 0.028 |
0.972 ± 0.014 | 3.267 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
