
Hypoxylon sp. CI-4A
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Xylariomycetidae; Xylariales; Hypoxylaceae; Hypoxylon; unclassified Hypoxylon
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
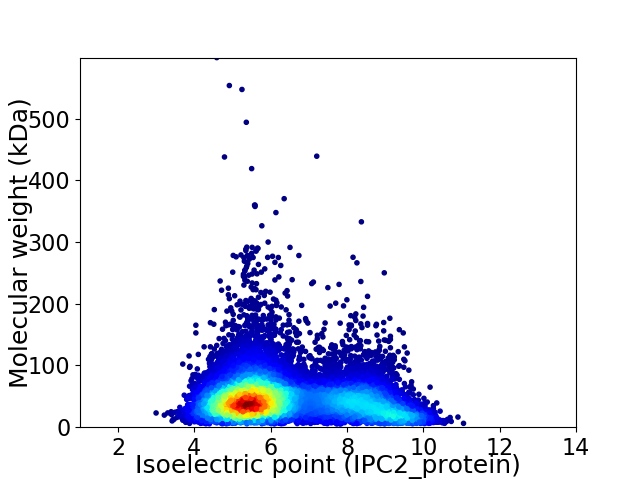
Virtual 2D-PAGE plot for 11701 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y2VNZ9|A0A1Y2VNZ9_9PEZI Aminotran_1_2 domain-containing protein OS=Hypoxylon sp. CI-4A OX=1001833 GN=M426DRAFT_258948 PE=4 SV=1
MM1 pKa = 6.95KK2 pKa = 8.92TWSSILPFVALLLPALIAADD22 pKa = 3.94CDD24 pKa = 3.96DD25 pKa = 4.16FTFTGQGGSDD35 pKa = 4.0GATEE39 pKa = 5.17PYY41 pKa = 9.76PSSFPVSASINCTTSLASQDD61 pKa = 2.94GGKK64 pKa = 9.76NDD66 pKa = 3.09KK67 pKa = 11.25CEE69 pKa = 3.87FHH71 pKa = 7.34HH72 pKa = 6.88YY73 pKa = 11.2NMGLVVHH80 pKa = 6.23PTINLTTINNSTRR93 pKa = 11.84DD94 pKa = 3.9DD95 pKa = 3.29IFALVKK101 pKa = 10.77DD102 pKa = 4.34KK103 pKa = 11.27ASSAADD109 pKa = 3.26TDD111 pKa = 4.36FNTTIVVNYY120 pKa = 7.41TAHH123 pKa = 5.16TQEE126 pKa = 4.77LSVGQAGHH134 pKa = 6.48VGFTPYY140 pKa = 10.1LRR142 pKa = 11.84CWEE145 pKa = 4.42GVVSDD150 pKa = 5.45CDD152 pKa = 5.59DD153 pKa = 5.71DD154 pKa = 7.4DD155 pKa = 7.46DD156 pKa = 7.5DD157 pKa = 7.47DD158 pKa = 6.2DD159 pKa = 5.92VDD161 pKa = 3.86VDD163 pKa = 3.17EE164 pKa = 4.86DD165 pKa = 4.08TAVRR169 pKa = 11.84VCGLIWLGGTEE180 pKa = 4.2GLSPGLQEE188 pKa = 4.64YY189 pKa = 10.35RR190 pKa = 11.84GEE192 pKa = 4.23EE193 pKa = 3.94NFVSSNVDD201 pKa = 2.59EE202 pKa = 5.05DD203 pKa = 4.87ANGDD207 pKa = 3.86LQPSYY212 pKa = 11.51DD213 pKa = 3.74SVAGQAIVTQGDD225 pKa = 3.24DD226 pKa = 3.15GAAINEE232 pKa = 4.08NHH234 pKa = 6.17EE235 pKa = 4.39VQQ237 pKa = 4.12
MM1 pKa = 6.95KK2 pKa = 8.92TWSSILPFVALLLPALIAADD22 pKa = 3.94CDD24 pKa = 3.96DD25 pKa = 4.16FTFTGQGGSDD35 pKa = 4.0GATEE39 pKa = 5.17PYY41 pKa = 9.76PSSFPVSASINCTTSLASQDD61 pKa = 2.94GGKK64 pKa = 9.76NDD66 pKa = 3.09KK67 pKa = 11.25CEE69 pKa = 3.87FHH71 pKa = 7.34HH72 pKa = 6.88YY73 pKa = 11.2NMGLVVHH80 pKa = 6.23PTINLTTINNSTRR93 pKa = 11.84DD94 pKa = 3.9DD95 pKa = 3.29IFALVKK101 pKa = 10.77DD102 pKa = 4.34KK103 pKa = 11.27ASSAADD109 pKa = 3.26TDD111 pKa = 4.36FNTTIVVNYY120 pKa = 7.41TAHH123 pKa = 5.16TQEE126 pKa = 4.77LSVGQAGHH134 pKa = 6.48VGFTPYY140 pKa = 10.1LRR142 pKa = 11.84CWEE145 pKa = 4.42GVVSDD150 pKa = 5.45CDD152 pKa = 5.59DD153 pKa = 5.71DD154 pKa = 7.4DD155 pKa = 7.46DD156 pKa = 7.5DD157 pKa = 7.47DD158 pKa = 6.2DD159 pKa = 5.92VDD161 pKa = 3.86VDD163 pKa = 3.17EE164 pKa = 4.86DD165 pKa = 4.08TAVRR169 pKa = 11.84VCGLIWLGGTEE180 pKa = 4.2GLSPGLQEE188 pKa = 4.64YY189 pKa = 10.35RR190 pKa = 11.84GEE192 pKa = 4.23EE193 pKa = 3.94NFVSSNVDD201 pKa = 2.59EE202 pKa = 5.05DD203 pKa = 4.87ANGDD207 pKa = 3.86LQPSYY212 pKa = 11.51DD213 pKa = 3.74SVAGQAIVTQGDD225 pKa = 3.24DD226 pKa = 3.15GAAINEE232 pKa = 4.08NHH234 pKa = 6.17EE235 pKa = 4.39VQQ237 pKa = 4.12
Molecular weight: 25.3 kDa
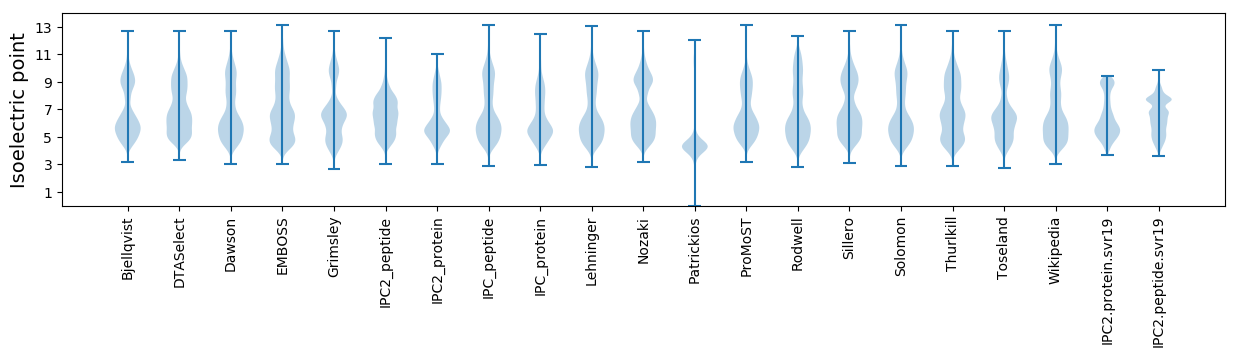
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y2W5E6|A0A1Y2W5E6_9PEZI NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit OS=Hypoxylon sp. CI-4A OX=1001833 GN=M426DRAFT_320301 PE=3 SV=1
LL1 pKa = 7.12LHH3 pKa = 7.12LLLFLLFFLLFFLLLLLPLLPLPPLLLSHH32 pKa = 6.97NLPSLLLTRR41 pKa = 11.84PRR43 pKa = 11.84PPMRR47 pKa = 11.84MMQISTNASVRR58 pKa = 11.84GAIAAQPRR66 pKa = 11.84GGVAALQLALANAPAPRR83 pKa = 11.84VGPVVLAPQLDD94 pKa = 3.73VAAFQTQRR102 pKa = 11.84RR103 pKa = 11.84RR104 pKa = 11.84RR105 pKa = 11.84LDD107 pKa = 3.56PDD109 pKa = 3.31QPPLVVVVPRR119 pKa = 11.84RR120 pKa = 11.84LRR122 pKa = 11.84GRR124 pKa = 11.84VLALAVRR131 pKa = 11.84AGAGRR136 pKa = 11.84RR137 pKa = 11.84RR138 pKa = 11.84RR139 pKa = 11.84RR140 pKa = 11.84RR141 pKa = 11.84RR142 pKa = 11.84VRR144 pKa = 11.84HH145 pKa = 5.6GGWW148 pKa = 2.85
LL1 pKa = 7.12LHH3 pKa = 7.12LLLFLLFFLLFFLLLLLPLLPLPPLLLSHH32 pKa = 6.97NLPSLLLTRR41 pKa = 11.84PRR43 pKa = 11.84PPMRR47 pKa = 11.84MMQISTNASVRR58 pKa = 11.84GAIAAQPRR66 pKa = 11.84GGVAALQLALANAPAPRR83 pKa = 11.84VGPVVLAPQLDD94 pKa = 3.73VAAFQTQRR102 pKa = 11.84RR103 pKa = 11.84RR104 pKa = 11.84RR105 pKa = 11.84LDD107 pKa = 3.56PDD109 pKa = 3.31QPPLVVVVPRR119 pKa = 11.84RR120 pKa = 11.84LRR122 pKa = 11.84GRR124 pKa = 11.84VLALAVRR131 pKa = 11.84AGAGRR136 pKa = 11.84RR137 pKa = 11.84RR138 pKa = 11.84RR139 pKa = 11.84RR140 pKa = 11.84RR141 pKa = 11.84RR142 pKa = 11.84VRR144 pKa = 11.84HH145 pKa = 5.6GGWW148 pKa = 2.85
Molecular weight: 16.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5364672 |
50 |
5457 |
458.5 |
50.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.402 ± 0.025 | 1.174 ± 0.008 |
5.862 ± 0.015 | 6.194 ± 0.02 |
3.753 ± 0.014 | 6.909 ± 0.023 |
2.334 ± 0.01 | 5.065 ± 0.014 |
4.88 ± 0.022 | 8.783 ± 0.024 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.101 ± 0.008 | 3.827 ± 0.011 |
6.041 ± 0.022 | 3.928 ± 0.015 |
6.041 ± 0.02 | 8.227 ± 0.025 |
6.016 ± 0.015 | 6.101 ± 0.017 |
1.494 ± 0.008 | 2.865 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
