
Acremonium chrysogenum (strain ATCC 11550 / CBS 779.69 / DSM 880 / JCM 23072 / IMI 49137)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Hypocreomycetidae; Hypocreales; Hypocreales incertae sedis; Acremonium; Acremonium chrysogenum
Average proteome isoelectric point is 6.46
Get precalculated fractions of proteins
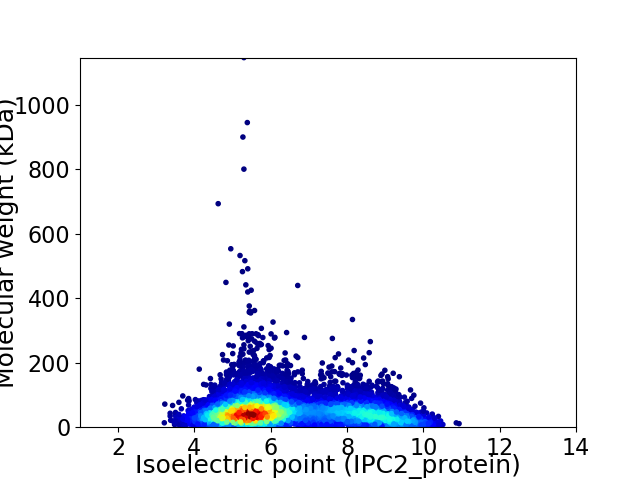
Virtual 2D-PAGE plot for 8899 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A086T0U6|A0A086T0U6_ACRC1 F-box domain-containing protein OS=Acremonium chrysogenum (strain ATCC 11550 / CBS 779.69 / DSM 880 / JCM 23072 / IMI 49137) OX=857340 GN=ACRE_062610 PE=4 SV=1
MM1 pKa = 7.78LDD3 pKa = 3.64KK4 pKa = 11.05SFPLNALSLLLLGGQASATYY24 pKa = 9.59LYY26 pKa = 10.02VSSYY30 pKa = 11.08SGIITTLKK38 pKa = 10.74LNGSGSEE45 pKa = 4.13TTLEE49 pKa = 4.03AVSSNDD55 pKa = 2.97EE56 pKa = 4.4CGGSASWLTLDD67 pKa = 4.13PSNGVIYY74 pKa = 9.13CTDD77 pKa = 3.78EE78 pKa = 4.0AWAGPPGAITSFATHH93 pKa = 6.84EE94 pKa = 4.7DD95 pKa = 3.62GSLSVLDD102 pKa = 3.86TQEE105 pKa = 4.02TFVGGVSVVQYY116 pKa = 11.28GDD118 pKa = 3.45NNNGLGVAYY127 pKa = 10.42YY128 pKa = 10.28GGSAFTTWDD137 pKa = 3.27ATDD140 pKa = 3.84PAALDD145 pKa = 3.75LVDD148 pKa = 4.25EE149 pKa = 5.06FVFEE153 pKa = 4.72LEE155 pKa = 4.39EE156 pKa = 4.63PGANPDD162 pKa = 3.53RR163 pKa = 11.84QEE165 pKa = 4.26APHH168 pKa = 6.25PHH170 pKa = 5.73QAALDD175 pKa = 3.89PSGEE179 pKa = 4.26FILVPDD185 pKa = 4.75LGADD189 pKa = 3.89LVRR192 pKa = 11.84TYY194 pKa = 10.72RR195 pKa = 11.84VAEE198 pKa = 4.06GDD200 pKa = 3.82FAVTPSGDD208 pKa = 3.67LEE210 pKa = 4.4LPAGSGPRR218 pKa = 11.84HH219 pKa = 5.56LTFAVHH225 pKa = 7.23GDD227 pKa = 3.41TTYY230 pKa = 10.14MYY232 pKa = 11.14LVAEE236 pKa = 4.65LSNTIFGYY244 pKa = 7.68TVEE247 pKa = 4.39YY248 pKa = 9.87SSSGIDD254 pKa = 3.19FKK256 pKa = 11.19KK257 pKa = 9.8IYY259 pKa = 10.28EE260 pKa = 4.44SGTHH264 pKa = 6.01GEE266 pKa = 4.25GVEE269 pKa = 4.28VPEE272 pKa = 4.68GASAAEE278 pKa = 3.91IVVTSDD284 pKa = 2.71QKK286 pKa = 11.32YY287 pKa = 10.39LIVSSRR293 pKa = 11.84NEE295 pKa = 4.08NIFEE299 pKa = 4.33IPSFDD304 pKa = 3.96PDD306 pKa = 3.32SDD308 pKa = 3.67EE309 pKa = 4.94TIVSDD314 pKa = 3.43TLINFSIDD322 pKa = 3.35PATGALSQLQEE333 pKa = 4.22VPSGGRR339 pKa = 11.84VPRR342 pKa = 11.84HH343 pKa = 5.47FSLNADD349 pKa = 3.71EE350 pKa = 4.55TQVAVVQQSDD360 pKa = 3.53GRR362 pKa = 11.84VVLIEE367 pKa = 4.46RR368 pKa = 11.84DD369 pKa = 3.84PEE371 pKa = 4.71SGLLGDD377 pKa = 5.03FVAAADD383 pKa = 4.3VEE385 pKa = 4.79GQVNTVIFNEE395 pKa = 4.77LII397 pKa = 3.9
MM1 pKa = 7.78LDD3 pKa = 3.64KK4 pKa = 11.05SFPLNALSLLLLGGQASATYY24 pKa = 9.59LYY26 pKa = 10.02VSSYY30 pKa = 11.08SGIITTLKK38 pKa = 10.74LNGSGSEE45 pKa = 4.13TTLEE49 pKa = 4.03AVSSNDD55 pKa = 2.97EE56 pKa = 4.4CGGSASWLTLDD67 pKa = 4.13PSNGVIYY74 pKa = 9.13CTDD77 pKa = 3.78EE78 pKa = 4.0AWAGPPGAITSFATHH93 pKa = 6.84EE94 pKa = 4.7DD95 pKa = 3.62GSLSVLDD102 pKa = 3.86TQEE105 pKa = 4.02TFVGGVSVVQYY116 pKa = 11.28GDD118 pKa = 3.45NNNGLGVAYY127 pKa = 10.42YY128 pKa = 10.28GGSAFTTWDD137 pKa = 3.27ATDD140 pKa = 3.84PAALDD145 pKa = 3.75LVDD148 pKa = 4.25EE149 pKa = 5.06FVFEE153 pKa = 4.72LEE155 pKa = 4.39EE156 pKa = 4.63PGANPDD162 pKa = 3.53RR163 pKa = 11.84QEE165 pKa = 4.26APHH168 pKa = 6.25PHH170 pKa = 5.73QAALDD175 pKa = 3.89PSGEE179 pKa = 4.26FILVPDD185 pKa = 4.75LGADD189 pKa = 3.89LVRR192 pKa = 11.84TYY194 pKa = 10.72RR195 pKa = 11.84VAEE198 pKa = 4.06GDD200 pKa = 3.82FAVTPSGDD208 pKa = 3.67LEE210 pKa = 4.4LPAGSGPRR218 pKa = 11.84HH219 pKa = 5.56LTFAVHH225 pKa = 7.23GDD227 pKa = 3.41TTYY230 pKa = 10.14MYY232 pKa = 11.14LVAEE236 pKa = 4.65LSNTIFGYY244 pKa = 7.68TVEE247 pKa = 4.39YY248 pKa = 9.87SSSGIDD254 pKa = 3.19FKK256 pKa = 11.19KK257 pKa = 9.8IYY259 pKa = 10.28EE260 pKa = 4.44SGTHH264 pKa = 6.01GEE266 pKa = 4.25GVEE269 pKa = 4.28VPEE272 pKa = 4.68GASAAEE278 pKa = 3.91IVVTSDD284 pKa = 2.71QKK286 pKa = 11.32YY287 pKa = 10.39LIVSSRR293 pKa = 11.84NEE295 pKa = 4.08NIFEE299 pKa = 4.33IPSFDD304 pKa = 3.96PDD306 pKa = 3.32SDD308 pKa = 3.67EE309 pKa = 4.94TIVSDD314 pKa = 3.43TLINFSIDD322 pKa = 3.35PATGALSQLQEE333 pKa = 4.22VPSGGRR339 pKa = 11.84VPRR342 pKa = 11.84HH343 pKa = 5.47FSLNADD349 pKa = 3.71EE350 pKa = 4.55TQVAVVQQSDD360 pKa = 3.53GRR362 pKa = 11.84VVLIEE367 pKa = 4.46RR368 pKa = 11.84DD369 pKa = 3.84PEE371 pKa = 4.71SGLLGDD377 pKa = 5.03FVAAADD383 pKa = 4.3VEE385 pKa = 4.79GQVNTVIFNEE395 pKa = 4.77LII397 pKa = 3.9
Molecular weight: 42.02 kDa
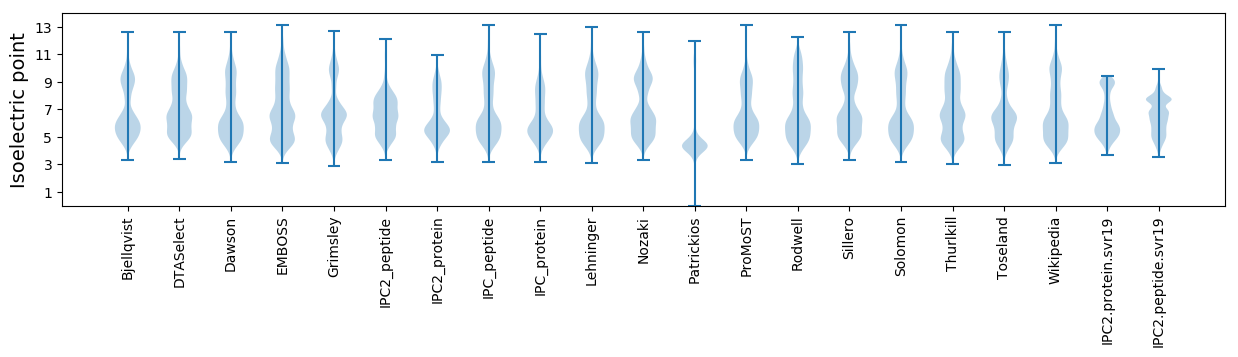
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A086SX64|A0A086SX64_ACRC1 Uncharacterized protein OS=Acremonium chrysogenum (strain ATCC 11550 / CBS 779.69 / DSM 880 / JCM 23072 / IMI 49137) OX=857340 GN=ACRE_075940 PE=4 SV=1
MM1 pKa = 7.26NALTRR6 pKa = 11.84SFTRR10 pKa = 11.84LAVRR14 pKa = 11.84PAGAATKK21 pKa = 10.25SSRR24 pKa = 11.84TFTTLSPHH32 pKa = 6.74RR33 pKa = 11.84PTVLPSAITTRR44 pKa = 11.84LPTSFTPSAGAGADD58 pKa = 3.88LVPTAAITRR67 pKa = 11.84HH68 pKa = 6.13PALQAIRR75 pKa = 11.84CGPRR79 pKa = 11.84NTMNGATRR87 pKa = 11.84MVQKK91 pKa = 10.21RR92 pKa = 11.84RR93 pKa = 11.84SGFLVRR99 pKa = 11.84KK100 pKa = 9.94RR101 pKa = 11.84SRR103 pKa = 11.84TGRR106 pKa = 11.84KK107 pKa = 8.1ILLRR111 pKa = 11.84RR112 pKa = 11.84RR113 pKa = 11.84MKK115 pKa = 10.2GRR117 pKa = 11.84RR118 pKa = 11.84EE119 pKa = 3.56IAQQ122 pKa = 3.29
MM1 pKa = 7.26NALTRR6 pKa = 11.84SFTRR10 pKa = 11.84LAVRR14 pKa = 11.84PAGAATKK21 pKa = 10.25SSRR24 pKa = 11.84TFTTLSPHH32 pKa = 6.74RR33 pKa = 11.84PTVLPSAITTRR44 pKa = 11.84LPTSFTPSAGAGADD58 pKa = 3.88LVPTAAITRR67 pKa = 11.84HH68 pKa = 6.13PALQAIRR75 pKa = 11.84CGPRR79 pKa = 11.84NTMNGATRR87 pKa = 11.84MVQKK91 pKa = 10.21RR92 pKa = 11.84RR93 pKa = 11.84SGFLVRR99 pKa = 11.84KK100 pKa = 9.94RR101 pKa = 11.84SRR103 pKa = 11.84TGRR106 pKa = 11.84KK107 pKa = 8.1ILLRR111 pKa = 11.84RR112 pKa = 11.84RR113 pKa = 11.84MKK115 pKa = 10.2GRR117 pKa = 11.84RR118 pKa = 11.84EE119 pKa = 3.56IAQQ122 pKa = 3.29
Molecular weight: 13.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4365916 |
25 |
10359 |
490.6 |
54.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.127 ± 0.024 | 1.162 ± 0.01 |
6.094 ± 0.017 | 6.351 ± 0.026 |
3.459 ± 0.015 | 7.328 ± 0.026 |
2.426 ± 0.01 | 4.364 ± 0.017 |
4.62 ± 0.025 | 8.637 ± 0.029 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.23 ± 0.009 | 3.303 ± 0.014 |
6.408 ± 0.029 | 3.924 ± 0.018 |
6.665 ± 0.025 | 7.948 ± 0.029 |
5.738 ± 0.017 | 6.191 ± 0.023 |
1.441 ± 0.009 | 2.582 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
