
Fusarium poae
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Hypocreomycetidae; Hypocreales; Nectriaceae; Fusarium; Fusarium sambucinum species complex
Average proteome isoelectric point is 6.39
Get precalculated fractions of proteins
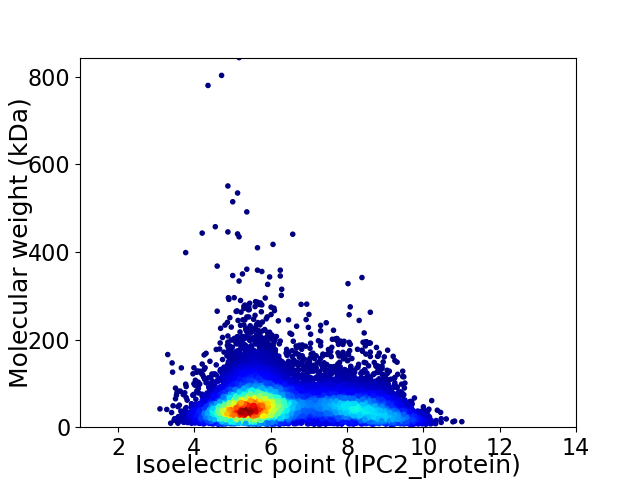
Virtual 2D-PAGE plot for 14048 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B8AFE5|A0A1B8AFE5_FUSPO Importin N-terminal domain-containing protein OS=Fusarium poae OX=36050 GN=FPOA_10887 PE=4 SV=1
MM1 pKa = 7.91RR2 pKa = 11.84ISASLGVLVASGLLGHH18 pKa = 6.35SASQATDD25 pKa = 3.36YY26 pKa = 11.58SNTNGSPFGNQPDD39 pKa = 4.04GSGNAIPGNNGGVSNTLPYY58 pKa = 9.96PSNAVGSIPGSLPSGVGPVGPVFPGDD84 pKa = 3.79GSNGLPPTGPVGGPEE99 pKa = 4.14NLPGSASPGGGSNGPYY115 pKa = 9.83PNGPGGIPGNLPGSGSPADD134 pKa = 4.54DD135 pKa = 4.66ANGLPPNGPTGGPGNVPGSIPGSSSSNGNGNFPGAGSVIIPGSFPTSIPQNVPSSPDD192 pKa = 3.3SPLSGASDD200 pKa = 3.32IPTCPFRR207 pKa = 11.84STKK210 pKa = 9.61TVVVTIYY217 pKa = 8.73PTDD220 pKa = 3.62TEE222 pKa = 4.56SEE224 pKa = 4.58SGFHH228 pKa = 6.49WPDD231 pKa = 3.72DD232 pKa = 4.07SDD234 pKa = 4.57SDD236 pKa = 4.0GSYY239 pKa = 10.17TPTPMPTKK247 pKa = 10.2PPGITSLSPNSGVPIFTTLTLDD269 pKa = 2.37IWGPVGSPSNSGSGSNGDD287 pKa = 3.32TSTSNGNQPSDD298 pKa = 2.99GSTPEE303 pKa = 4.85DD304 pKa = 3.45GSGNNGGNDD313 pKa = 3.21SHH315 pKa = 7.39GGNGPDD321 pKa = 4.35SNGGDD326 pKa = 3.88DD327 pKa = 3.84SHH329 pKa = 7.83NSSDD333 pKa = 4.23SPFFPPTPGPNGAPQYY349 pKa = 10.86SDD351 pKa = 3.3STSEE355 pKa = 4.1SLSVGNPGASGINTLAPTPLQTPEE379 pKa = 4.4FPNGGIPGTQPQFPSAGPAQEE400 pKa = 4.7ASGSLPVTTQAGAEE414 pKa = 4.39GQGEE418 pKa = 4.27TSPWFTNVPEE428 pKa = 4.72GNSAGQPTSPYY439 pKa = 9.19ITITGQDD446 pKa = 3.76GLPTVISSDD455 pKa = 3.69GNVVSGDD462 pKa = 3.76AQPPAVPGSPQNGNSPEE479 pKa = 4.16LPSGVSSGFPIPPPIFTGIPGSQNPSNSPAGSLPGSGPDD518 pKa = 4.34AGVTTCATFTITGPNGLPTVVDD540 pKa = 4.05STWVVPLSTPSFEE553 pKa = 5.46ASSQLSATFDD563 pKa = 3.5PQAPLSSISSQLGLPTIPASGSPEE587 pKa = 4.32DD588 pKa = 4.12GSGGSAAIASTSFTMIGADD607 pKa = 3.58GSPTVVYY614 pKa = 7.28TTWNIPAATISTGDD628 pKa = 3.7PGDD631 pKa = 3.92ASTAASSGGFMTITGLPPFGSSGLGGQITTVPGPDD666 pKa = 3.51DD667 pKa = 3.38VAQITTCTSYY677 pKa = 10.58TVIGADD683 pKa = 3.72GLPTVIDD690 pKa = 3.71STFVIPGPVNTDD702 pKa = 2.53ITVGGDD708 pKa = 3.25PEE710 pKa = 4.64LPSGASDD717 pKa = 3.72ALPNGVTSPATFQVTNTPEE736 pKa = 3.94IPPFNPTAATGSGGFGNNGITTCTSYY762 pKa = 11.27TMIGSDD768 pKa = 3.94GLPTVVDD775 pKa = 4.1TTWVIPGSANTQSEE789 pKa = 4.72LPGNPSFVSDD799 pKa = 4.17SLPTGLPSGIPGPWASSAGIPSDD822 pKa = 3.57GDD824 pKa = 3.24QGNVPGATTCITYY837 pKa = 7.52TTIGQDD843 pKa = 3.73GLPTIVDD850 pKa = 3.6TTFVVPVATATPSGTGLILPSTNGDD875 pKa = 3.51GQTGLSPNPTGAYY888 pKa = 5.9TTTTTAAILGSDD900 pKa = 4.02GNATPTVQTIVFSDD914 pKa = 3.83SSALGVSTSAPQGATSGIDD933 pKa = 3.22PSGISGPVFTSRR945 pKa = 11.84DD946 pKa = 3.66LNSPFTDD953 pKa = 3.45TPSLNGYY960 pKa = 10.53DD961 pKa = 3.44NGIPGASMSAILTDD975 pKa = 4.24GSAAGEE981 pKa = 4.44GTSVTGTTTGTLTWTVTSITNPPGAPVFSNGASQPPFVPDD1021 pKa = 4.39PSNGAPGGPAQPAYY1035 pKa = 10.46GSLATEE1041 pKa = 4.07STLWPLSAVQTSTWTNVIKK1060 pKa = 10.92AEE1062 pKa = 4.24TTSYY1066 pKa = 9.43TFNYY1070 pKa = 9.59PLTTLATVNVPMRR1083 pKa = 11.84RR1084 pKa = 11.84LARR1087 pKa = 11.84RR1088 pKa = 11.84QSMTAWSNSTTSLSSATSASTVTTSEE1114 pKa = 3.86ASSPTVCPSGGSIGNTTIDD1133 pKa = 3.95FDD1135 pKa = 4.04NSKK1138 pKa = 10.22PGPLFNPVEE1147 pKa = 4.53NIWFSGGFLIAPPTSQQSQPYY1168 pKa = 8.41IPSSGGQLVEE1178 pKa = 4.92FVPPALSNTTTTISGDD1194 pKa = 3.28VAQIGMGPHH1203 pKa = 6.48AASPCFRR1210 pKa = 11.84FDD1212 pKa = 3.8FFGANLGCDD1221 pKa = 3.22ARR1223 pKa = 11.84GDD1225 pKa = 4.04EE1226 pKa = 4.75KK1227 pKa = 10.66WCQFDD1232 pKa = 3.21ISAYY1236 pKa = 9.13RR1237 pKa = 11.84WNEE1240 pKa = 3.59TSSTEE1245 pKa = 4.22EE1246 pKa = 3.76SIAWSEE1252 pKa = 4.39TKK1254 pKa = 9.88QVPACSKK1261 pKa = 9.97FSEE1264 pKa = 4.56GGYY1267 pKa = 10.31EE1268 pKa = 4.07LTRR1271 pKa = 11.84VDD1273 pKa = 3.82LDD1275 pKa = 3.85GYY1277 pKa = 11.04KK1278 pKa = 10.5DD1279 pKa = 3.72LSSVLITLRR1288 pKa = 11.84VSSDD1292 pKa = 2.96LRR1294 pKa = 11.84VWWGDD1299 pKa = 3.28DD1300 pKa = 3.8FRR1302 pKa = 11.84VGWSDD1307 pKa = 3.36NSCIAATCRR1316 pKa = 11.84ANAPSQFVKK1325 pKa = 10.72RR1326 pKa = 11.84EE1327 pKa = 4.04TVISALRR1334 pKa = 11.84QGVYY1338 pKa = 8.97RR1339 pKa = 11.84WTPYY1343 pKa = 9.24EE1344 pKa = 4.38LKK1346 pKa = 10.64RR1347 pKa = 11.84LDD1349 pKa = 5.16DD1350 pKa = 3.85SLVWEE1355 pKa = 4.61SANN1358 pKa = 3.76
MM1 pKa = 7.91RR2 pKa = 11.84ISASLGVLVASGLLGHH18 pKa = 6.35SASQATDD25 pKa = 3.36YY26 pKa = 11.58SNTNGSPFGNQPDD39 pKa = 4.04GSGNAIPGNNGGVSNTLPYY58 pKa = 9.96PSNAVGSIPGSLPSGVGPVGPVFPGDD84 pKa = 3.79GSNGLPPTGPVGGPEE99 pKa = 4.14NLPGSASPGGGSNGPYY115 pKa = 9.83PNGPGGIPGNLPGSGSPADD134 pKa = 4.54DD135 pKa = 4.66ANGLPPNGPTGGPGNVPGSIPGSSSSNGNGNFPGAGSVIIPGSFPTSIPQNVPSSPDD192 pKa = 3.3SPLSGASDD200 pKa = 3.32IPTCPFRR207 pKa = 11.84STKK210 pKa = 9.61TVVVTIYY217 pKa = 8.73PTDD220 pKa = 3.62TEE222 pKa = 4.56SEE224 pKa = 4.58SGFHH228 pKa = 6.49WPDD231 pKa = 3.72DD232 pKa = 4.07SDD234 pKa = 4.57SDD236 pKa = 4.0GSYY239 pKa = 10.17TPTPMPTKK247 pKa = 10.2PPGITSLSPNSGVPIFTTLTLDD269 pKa = 2.37IWGPVGSPSNSGSGSNGDD287 pKa = 3.32TSTSNGNQPSDD298 pKa = 2.99GSTPEE303 pKa = 4.85DD304 pKa = 3.45GSGNNGGNDD313 pKa = 3.21SHH315 pKa = 7.39GGNGPDD321 pKa = 4.35SNGGDD326 pKa = 3.88DD327 pKa = 3.84SHH329 pKa = 7.83NSSDD333 pKa = 4.23SPFFPPTPGPNGAPQYY349 pKa = 10.86SDD351 pKa = 3.3STSEE355 pKa = 4.1SLSVGNPGASGINTLAPTPLQTPEE379 pKa = 4.4FPNGGIPGTQPQFPSAGPAQEE400 pKa = 4.7ASGSLPVTTQAGAEE414 pKa = 4.39GQGEE418 pKa = 4.27TSPWFTNVPEE428 pKa = 4.72GNSAGQPTSPYY439 pKa = 9.19ITITGQDD446 pKa = 3.76GLPTVISSDD455 pKa = 3.69GNVVSGDD462 pKa = 3.76AQPPAVPGSPQNGNSPEE479 pKa = 4.16LPSGVSSGFPIPPPIFTGIPGSQNPSNSPAGSLPGSGPDD518 pKa = 4.34AGVTTCATFTITGPNGLPTVVDD540 pKa = 4.05STWVVPLSTPSFEE553 pKa = 5.46ASSQLSATFDD563 pKa = 3.5PQAPLSSISSQLGLPTIPASGSPEE587 pKa = 4.32DD588 pKa = 4.12GSGGSAAIASTSFTMIGADD607 pKa = 3.58GSPTVVYY614 pKa = 7.28TTWNIPAATISTGDD628 pKa = 3.7PGDD631 pKa = 3.92ASTAASSGGFMTITGLPPFGSSGLGGQITTVPGPDD666 pKa = 3.51DD667 pKa = 3.38VAQITTCTSYY677 pKa = 10.58TVIGADD683 pKa = 3.72GLPTVIDD690 pKa = 3.71STFVIPGPVNTDD702 pKa = 2.53ITVGGDD708 pKa = 3.25PEE710 pKa = 4.64LPSGASDD717 pKa = 3.72ALPNGVTSPATFQVTNTPEE736 pKa = 3.94IPPFNPTAATGSGGFGNNGITTCTSYY762 pKa = 11.27TMIGSDD768 pKa = 3.94GLPTVVDD775 pKa = 4.1TTWVIPGSANTQSEE789 pKa = 4.72LPGNPSFVSDD799 pKa = 4.17SLPTGLPSGIPGPWASSAGIPSDD822 pKa = 3.57GDD824 pKa = 3.24QGNVPGATTCITYY837 pKa = 7.52TTIGQDD843 pKa = 3.73GLPTIVDD850 pKa = 3.6TTFVVPVATATPSGTGLILPSTNGDD875 pKa = 3.51GQTGLSPNPTGAYY888 pKa = 5.9TTTTTAAILGSDD900 pKa = 4.02GNATPTVQTIVFSDD914 pKa = 3.83SSALGVSTSAPQGATSGIDD933 pKa = 3.22PSGISGPVFTSRR945 pKa = 11.84DD946 pKa = 3.66LNSPFTDD953 pKa = 3.45TPSLNGYY960 pKa = 10.53DD961 pKa = 3.44NGIPGASMSAILTDD975 pKa = 4.24GSAAGEE981 pKa = 4.44GTSVTGTTTGTLTWTVTSITNPPGAPVFSNGASQPPFVPDD1021 pKa = 4.39PSNGAPGGPAQPAYY1035 pKa = 10.46GSLATEE1041 pKa = 4.07STLWPLSAVQTSTWTNVIKK1060 pKa = 10.92AEE1062 pKa = 4.24TTSYY1066 pKa = 9.43TFNYY1070 pKa = 9.59PLTTLATVNVPMRR1083 pKa = 11.84RR1084 pKa = 11.84LARR1087 pKa = 11.84RR1088 pKa = 11.84QSMTAWSNSTTSLSSATSASTVTTSEE1114 pKa = 3.86ASSPTVCPSGGSIGNTTIDD1133 pKa = 3.95FDD1135 pKa = 4.04NSKK1138 pKa = 10.22PGPLFNPVEE1147 pKa = 4.53NIWFSGGFLIAPPTSQQSQPYY1168 pKa = 8.41IPSSGGQLVEE1178 pKa = 4.92FVPPALSNTTTTISGDD1194 pKa = 3.28VAQIGMGPHH1203 pKa = 6.48AASPCFRR1210 pKa = 11.84FDD1212 pKa = 3.8FFGANLGCDD1221 pKa = 3.22ARR1223 pKa = 11.84GDD1225 pKa = 4.04EE1226 pKa = 4.75KK1227 pKa = 10.66WCQFDD1232 pKa = 3.21ISAYY1236 pKa = 9.13RR1237 pKa = 11.84WNEE1240 pKa = 3.59TSSTEE1245 pKa = 4.22EE1246 pKa = 3.76SIAWSEE1252 pKa = 4.39TKK1254 pKa = 9.88QVPACSKK1261 pKa = 9.97FSEE1264 pKa = 4.56GGYY1267 pKa = 10.31EE1268 pKa = 4.07LTRR1271 pKa = 11.84VDD1273 pKa = 3.82LDD1275 pKa = 3.85GYY1277 pKa = 11.04KK1278 pKa = 10.5DD1279 pKa = 3.72LSSVLITLRR1288 pKa = 11.84VSSDD1292 pKa = 2.96LRR1294 pKa = 11.84VWWGDD1299 pKa = 3.28DD1300 pKa = 3.8FRR1302 pKa = 11.84VGWSDD1307 pKa = 3.36NSCIAATCRR1316 pKa = 11.84ANAPSQFVKK1325 pKa = 10.72RR1326 pKa = 11.84EE1327 pKa = 4.04TVISALRR1334 pKa = 11.84QGVYY1338 pKa = 8.97RR1339 pKa = 11.84WTPYY1343 pKa = 9.24EE1344 pKa = 4.38LKK1346 pKa = 10.64RR1347 pKa = 11.84LDD1349 pKa = 5.16DD1350 pKa = 3.85SLVWEE1355 pKa = 4.61SANN1358 pKa = 3.76
Molecular weight: 135.46 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B8B8E8|A0A1B8B8E8_FUSPO Uncharacterized protein OS=Fusarium poae OX=36050 GN=FPOA_02941 PE=4 SV=1
MM1 pKa = 7.86PLTRR5 pKa = 11.84THH7 pKa = 6.34RR8 pKa = 11.84HH9 pKa = 4.3SAPRR13 pKa = 11.84RR14 pKa = 11.84SIFSTRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84APARR26 pKa = 11.84SNRR29 pKa = 11.84HH30 pKa = 4.32TVTTTTTTTTKK41 pKa = 9.91PRR43 pKa = 11.84RR44 pKa = 11.84GMFGGGSTARR54 pKa = 11.84RR55 pKa = 11.84THH57 pKa = 5.96GAAPVHH63 pKa = 5.2HH64 pKa = 6.81HH65 pKa = 5.26QRR67 pKa = 11.84KK68 pKa = 8.6PSMKK72 pKa = 9.93DD73 pKa = 3.0KK74 pKa = 11.38VSGALLKK81 pKa = 10.61IKK83 pKa = 10.52GSLTRR88 pKa = 11.84RR89 pKa = 11.84PGVKK93 pKa = 9.89AAGTRR98 pKa = 11.84RR99 pKa = 11.84MHH101 pKa = 5.65GTDD104 pKa = 2.95GRR106 pKa = 11.84GARR109 pKa = 11.84HH110 pKa = 5.73RR111 pKa = 11.84RR112 pKa = 11.84YY113 pKa = 10.37
MM1 pKa = 7.86PLTRR5 pKa = 11.84THH7 pKa = 6.34RR8 pKa = 11.84HH9 pKa = 4.3SAPRR13 pKa = 11.84RR14 pKa = 11.84SIFSTRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84APARR26 pKa = 11.84SNRR29 pKa = 11.84HH30 pKa = 4.32TVTTTTTTTTKK41 pKa = 9.91PRR43 pKa = 11.84RR44 pKa = 11.84GMFGGGSTARR54 pKa = 11.84RR55 pKa = 11.84THH57 pKa = 5.96GAAPVHH63 pKa = 5.2HH64 pKa = 6.81HH65 pKa = 5.26QRR67 pKa = 11.84KK68 pKa = 8.6PSMKK72 pKa = 9.93DD73 pKa = 3.0KK74 pKa = 11.38VSGALLKK81 pKa = 10.61IKK83 pKa = 10.52GSLTRR88 pKa = 11.84RR89 pKa = 11.84PGVKK93 pKa = 9.89AAGTRR98 pKa = 11.84RR99 pKa = 11.84MHH101 pKa = 5.65GTDD104 pKa = 2.95GRR106 pKa = 11.84GARR109 pKa = 11.84HH110 pKa = 5.73RR111 pKa = 11.84RR112 pKa = 11.84YY113 pKa = 10.37
Molecular weight: 12.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7061983 |
42 |
7658 |
502.7 |
55.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.139 ± 0.02 | 1.307 ± 0.008 |
6.003 ± 0.016 | 6.347 ± 0.022 |
3.724 ± 0.011 | 6.605 ± 0.018 |
2.446 ± 0.01 | 5.109 ± 0.012 |
5.146 ± 0.017 | 8.685 ± 0.023 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.219 ± 0.009 | 3.839 ± 0.01 |
5.834 ± 0.021 | 4.109 ± 0.016 |
6.079 ± 0.017 | 8.041 ± 0.023 |
5.997 ± 0.022 | 6.005 ± 0.015 |
1.578 ± 0.008 | 2.787 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
