
Capybara microvirus Cap1_SP_66
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.67
Get precalculated fractions of proteins
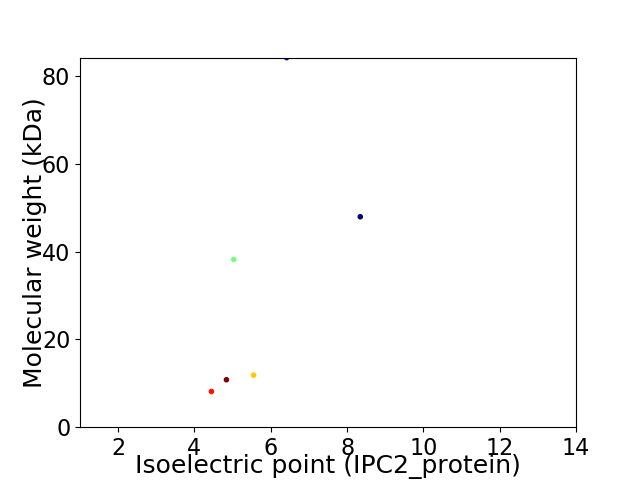
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
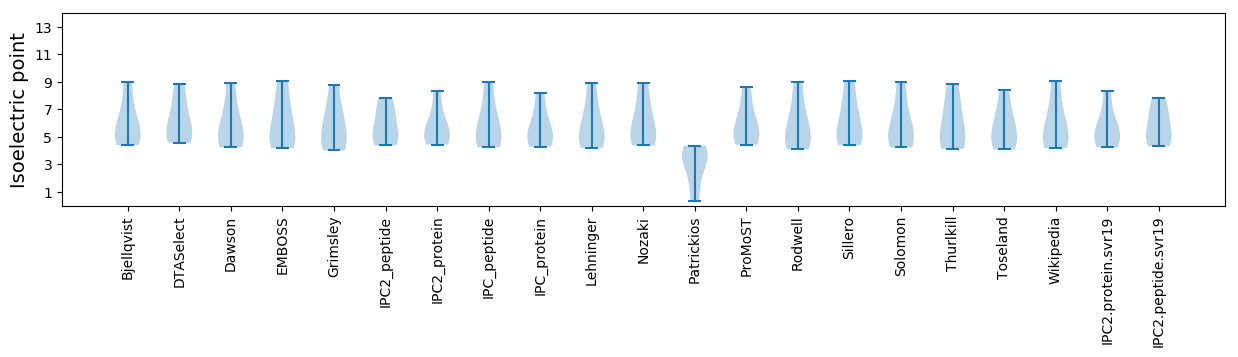
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W7J6|A0A4P8W7J6_9VIRU Uncharacterized protein OS=Capybara microvirus Cap1_SP_66 OX=2584788 PE=4 SV=1
MM1 pKa = 7.42TYY3 pKa = 10.53SIIALYY9 pKa = 10.25EE10 pKa = 4.11DD11 pKa = 4.41EE12 pKa = 5.43NEE14 pKa = 4.31CLHH17 pKa = 6.96QITLFNRR24 pKa = 11.84LCSLDD29 pKa = 3.45SALPLLKK36 pKa = 10.72NIFQTPSDD44 pKa = 4.38YY45 pKa = 10.55IACDD49 pKa = 3.32SLVLVDD55 pKa = 6.0CYY57 pKa = 10.94IVDD60 pKa = 3.83CEE62 pKa = 4.25GFRR65 pKa = 11.84VRR67 pKa = 11.84VSKK70 pKa = 11.17NN71 pKa = 2.82
MM1 pKa = 7.42TYY3 pKa = 10.53SIIALYY9 pKa = 10.25EE10 pKa = 4.11DD11 pKa = 4.41EE12 pKa = 5.43NEE14 pKa = 4.31CLHH17 pKa = 6.96QITLFNRR24 pKa = 11.84LCSLDD29 pKa = 3.45SALPLLKK36 pKa = 10.72NIFQTPSDD44 pKa = 4.38YY45 pKa = 10.55IACDD49 pKa = 3.32SLVLVDD55 pKa = 6.0CYY57 pKa = 10.94IVDD60 pKa = 3.83CEE62 pKa = 4.25GFRR65 pKa = 11.84VRR67 pKa = 11.84VSKK70 pKa = 11.17NN71 pKa = 2.82
Molecular weight: 8.14 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W3Y1|A0A4P8W3Y1_9VIRU Major capsid protein OS=Capybara microvirus Cap1_SP_66 OX=2584788 PE=3 SV=1
MM1 pKa = 7.27SCSNPTYY8 pKa = 10.47FFNKK12 pKa = 9.81KK13 pKa = 10.17FIQRR17 pKa = 11.84YY18 pKa = 7.74YY19 pKa = 10.03SLVDD23 pKa = 3.18GFHH26 pKa = 5.86YY27 pKa = 10.73HH28 pKa = 6.43GVDD31 pKa = 3.52YY32 pKa = 10.15PFKK35 pKa = 10.56PSVSDD40 pKa = 3.36VASIFDD46 pKa = 3.8EE47 pKa = 5.25FKK49 pKa = 10.87AGQTFDD55 pKa = 3.16FRR57 pKa = 11.84LYY59 pKa = 10.01LTGAYY64 pKa = 10.25DD65 pKa = 3.51GDD67 pKa = 4.44YY68 pKa = 10.59VVHH71 pKa = 5.96YY72 pKa = 8.16PINPFLVIPCHH83 pKa = 5.78NCPSCLHH90 pKa = 7.05DD91 pKa = 3.85KK92 pKa = 10.64QATKK96 pKa = 10.64SALIALEE103 pKa = 4.39SVGKK107 pKa = 9.33PSPLFVTLTYY117 pKa = 10.82ADD119 pKa = 3.9EE120 pKa = 5.31PINGVSVRR128 pKa = 11.84DD129 pKa = 3.44VQLFLKK135 pKa = 10.16RR136 pKa = 11.84LRR138 pKa = 11.84KK139 pKa = 8.74MFPQSKK145 pKa = 9.36IRR147 pKa = 11.84YY148 pKa = 7.82HH149 pKa = 5.9CTAEE153 pKa = 4.37YY154 pKa = 10.68GSHH157 pKa = 6.22TYY159 pKa = 9.92RR160 pKa = 11.84PHH162 pKa = 4.93YY163 pKa = 9.87HH164 pKa = 6.75LLIFGLPLEE173 pKa = 4.59SEE175 pKa = 4.4QFVEE179 pKa = 5.12SIKK182 pKa = 10.81SRR184 pKa = 11.84YY185 pKa = 8.37NVPNYY190 pKa = 9.3PVGYY194 pKa = 9.74GYY196 pKa = 11.25YY197 pKa = 9.43LDD199 pKa = 3.92YY200 pKa = 10.84SIRR203 pKa = 11.84QAWQLGITHH212 pKa = 6.19SRR214 pKa = 11.84VVLNPAVIRR223 pKa = 11.84YY224 pKa = 7.26VSKK227 pKa = 9.85YY228 pKa = 7.4TSKK231 pKa = 10.81QSDD234 pKa = 3.73VLLGKK239 pKa = 10.02NPPFMLQSTHH249 pKa = 6.21NGGLGASVLRR259 pKa = 11.84DD260 pKa = 3.25YY261 pKa = 10.96IINNNFDD268 pKa = 4.29SNCGISLNGRR278 pKa = 11.84YY279 pKa = 7.43FTIPSNLFNQRR290 pKa = 11.84VCLSINRR297 pKa = 11.84YY298 pKa = 8.4IGYY301 pKa = 9.69YY302 pKa = 8.22WLLVMKK308 pKa = 10.53ALDD311 pKa = 4.23HH312 pKa = 7.26LSLLPDD318 pKa = 4.27LPPIIMQAVHH328 pKa = 6.57SYY330 pKa = 9.89IVPRR334 pKa = 11.84GTNKK338 pKa = 10.11SLEE341 pKa = 3.95FLEE344 pKa = 4.75YY345 pKa = 10.48INDD348 pKa = 3.39EE349 pKa = 4.43TFPARR354 pKa = 11.84YY355 pKa = 8.83IPAVSARR362 pKa = 11.84FNDD365 pKa = 4.43DD366 pKa = 3.18YY367 pKa = 11.57AQKK370 pKa = 10.86LEE372 pKa = 3.83QFLKK376 pKa = 10.39IKK378 pKa = 10.67HH379 pKa = 5.37EE380 pKa = 5.0FIAKK384 pKa = 8.46RR385 pKa = 11.84TIPEE389 pKa = 3.91VPNQNFLQNYY399 pKa = 7.78RR400 pKa = 11.84AEE402 pKa = 4.16IQLNRR407 pKa = 11.84QKK409 pKa = 10.93EE410 pKa = 4.27KK411 pKa = 11.23EE412 pKa = 4.09KK413 pKa = 11.13LL414 pKa = 3.49
MM1 pKa = 7.27SCSNPTYY8 pKa = 10.47FFNKK12 pKa = 9.81KK13 pKa = 10.17FIQRR17 pKa = 11.84YY18 pKa = 7.74YY19 pKa = 10.03SLVDD23 pKa = 3.18GFHH26 pKa = 5.86YY27 pKa = 10.73HH28 pKa = 6.43GVDD31 pKa = 3.52YY32 pKa = 10.15PFKK35 pKa = 10.56PSVSDD40 pKa = 3.36VASIFDD46 pKa = 3.8EE47 pKa = 5.25FKK49 pKa = 10.87AGQTFDD55 pKa = 3.16FRR57 pKa = 11.84LYY59 pKa = 10.01LTGAYY64 pKa = 10.25DD65 pKa = 3.51GDD67 pKa = 4.44YY68 pKa = 10.59VVHH71 pKa = 5.96YY72 pKa = 8.16PINPFLVIPCHH83 pKa = 5.78NCPSCLHH90 pKa = 7.05DD91 pKa = 3.85KK92 pKa = 10.64QATKK96 pKa = 10.64SALIALEE103 pKa = 4.39SVGKK107 pKa = 9.33PSPLFVTLTYY117 pKa = 10.82ADD119 pKa = 3.9EE120 pKa = 5.31PINGVSVRR128 pKa = 11.84DD129 pKa = 3.44VQLFLKK135 pKa = 10.16RR136 pKa = 11.84LRR138 pKa = 11.84KK139 pKa = 8.74MFPQSKK145 pKa = 9.36IRR147 pKa = 11.84YY148 pKa = 7.82HH149 pKa = 5.9CTAEE153 pKa = 4.37YY154 pKa = 10.68GSHH157 pKa = 6.22TYY159 pKa = 9.92RR160 pKa = 11.84PHH162 pKa = 4.93YY163 pKa = 9.87HH164 pKa = 6.75LLIFGLPLEE173 pKa = 4.59SEE175 pKa = 4.4QFVEE179 pKa = 5.12SIKK182 pKa = 10.81SRR184 pKa = 11.84YY185 pKa = 8.37NVPNYY190 pKa = 9.3PVGYY194 pKa = 9.74GYY196 pKa = 11.25YY197 pKa = 9.43LDD199 pKa = 3.92YY200 pKa = 10.84SIRR203 pKa = 11.84QAWQLGITHH212 pKa = 6.19SRR214 pKa = 11.84VVLNPAVIRR223 pKa = 11.84YY224 pKa = 7.26VSKK227 pKa = 9.85YY228 pKa = 7.4TSKK231 pKa = 10.81QSDD234 pKa = 3.73VLLGKK239 pKa = 10.02NPPFMLQSTHH249 pKa = 6.21NGGLGASVLRR259 pKa = 11.84DD260 pKa = 3.25YY261 pKa = 10.96IINNNFDD268 pKa = 4.29SNCGISLNGRR278 pKa = 11.84YY279 pKa = 7.43FTIPSNLFNQRR290 pKa = 11.84VCLSINRR297 pKa = 11.84YY298 pKa = 8.4IGYY301 pKa = 9.69YY302 pKa = 8.22WLLVMKK308 pKa = 10.53ALDD311 pKa = 4.23HH312 pKa = 7.26LSLLPDD318 pKa = 4.27LPPIIMQAVHH328 pKa = 6.57SYY330 pKa = 9.89IVPRR334 pKa = 11.84GTNKK338 pKa = 10.11SLEE341 pKa = 3.95FLEE344 pKa = 4.75YY345 pKa = 10.48INDD348 pKa = 3.39EE349 pKa = 4.43TFPARR354 pKa = 11.84YY355 pKa = 8.83IPAVSARR362 pKa = 11.84FNDD365 pKa = 4.43DD366 pKa = 3.18YY367 pKa = 11.57AQKK370 pKa = 10.86LEE372 pKa = 3.83QFLKK376 pKa = 10.39IKK378 pKa = 10.67HH379 pKa = 5.37EE380 pKa = 5.0FIAKK384 pKa = 8.46RR385 pKa = 11.84TIPEE389 pKa = 3.91VPNQNFLQNYY399 pKa = 7.78RR400 pKa = 11.84AEE402 pKa = 4.16IQLNRR407 pKa = 11.84QKK409 pKa = 10.93EE410 pKa = 4.27KK411 pKa = 11.23EE412 pKa = 4.09KK413 pKa = 11.13LL414 pKa = 3.49
Molecular weight: 47.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1777 |
71 |
750 |
296.2 |
33.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.246 ± 0.693 | 1.463 ± 0.288 |
5.909 ± 0.371 | 4.333 ± 0.746 |
4.502 ± 1.022 | 5.459 ± 0.606 |
2.195 ± 0.493 | 6.078 ± 0.243 |
5.065 ± 0.413 | 9.06 ± 0.844 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.589 ± 0.42 | 6.64 ± 0.586 |
5.065 ± 0.597 | 4.952 ± 0.629 |
4.389 ± 0.271 | 7.822 ± 0.532 |
5.796 ± 0.654 | 5.965 ± 0.273 |
0.732 ± 0.193 | 5.74 ± 0.957 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
