
Vibrio phage fs2
Taxonomy: Viruses; Monodnaviria; Loebvirae; Hofneiviricota; Faserviricetes; Tubulavirales; Inoviridae; Saetivirus; Vibrio virus fs2
Average proteome isoelectric point is 6.91
Get precalculated fractions of proteins
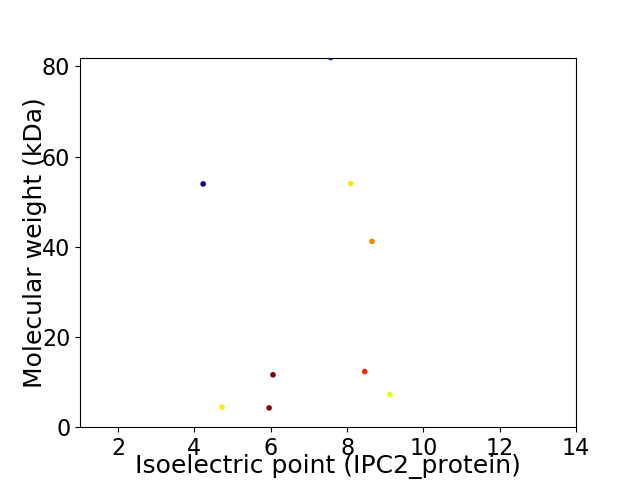
Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|O80262|O80262_9VIRU Coat protein D OS=Vibrio phage fs2 OX=83201 PE=3 SV=1
MM1 pKa = 7.24RR2 pKa = 11.84TVLTLFSTTLLLSISFSSFAYY23 pKa = 9.53YY24 pKa = 10.02QLPFWPDD31 pKa = 2.7KK32 pKa = 11.01TYY34 pKa = 9.5LTPEE38 pKa = 4.01AAAAAYY44 pKa = 10.57LDD46 pKa = 3.57ILGGGSCKK54 pKa = 9.81RR55 pKa = 11.84DD56 pKa = 2.96TRR58 pKa = 11.84NFIKK62 pKa = 10.75ASISSINTPHH72 pKa = 6.43IAYY75 pKa = 10.31RR76 pKa = 11.84IDD78 pKa = 4.32KK79 pKa = 10.47YY80 pKa = 11.39LDD82 pKa = 3.65DD83 pKa = 3.77QCIVLSSTGNASVTLNVVDD102 pKa = 4.11NCPDD106 pKa = 3.18GTSPDD111 pKa = 4.62LSTGMCKK118 pKa = 10.24PKK120 pKa = 10.12PDD122 pKa = 3.45TPQYY126 pKa = 10.28CGTSAMFEE134 pKa = 4.36DD135 pKa = 4.93VANLYY140 pKa = 7.91NACYY144 pKa = 8.22EE145 pKa = 4.14QNGILSYY152 pKa = 10.61TCDD155 pKa = 3.69EE156 pKa = 4.29STQALDD162 pKa = 3.92AKK164 pKa = 10.71CDD166 pKa = 3.69LTTSDD171 pKa = 3.0QCVIGRR177 pKa = 11.84PTWPDD182 pKa = 3.63CLDD185 pKa = 4.38KK186 pKa = 10.83PHH188 pKa = 6.6QPNDD192 pKa = 3.62PTNPLPPVGGFNPSPVNPSVPPSPVEE218 pKa = 3.9KK219 pKa = 10.15PDD221 pKa = 3.28VQEE224 pKa = 4.03PDD226 pKa = 3.1KK227 pKa = 11.5TEE229 pKa = 3.84TSDD232 pKa = 3.31TGVINAIKK240 pKa = 10.52NLNDD244 pKa = 4.59DD245 pKa = 4.33LNKK248 pKa = 10.79SNTDD252 pKa = 2.65IHH254 pKa = 8.08NDD256 pKa = 3.02MNNIFSTMNDD266 pKa = 3.02ALRR269 pKa = 11.84QLNSTNTAIGQSIVEE284 pKa = 4.13QMKK287 pKa = 10.24QDD289 pKa = 3.35AQIYY293 pKa = 9.46DD294 pKa = 3.74NKK296 pKa = 10.73KK297 pKa = 9.53LQQQIAANNINAINSQTKK315 pKa = 10.42SLLEE319 pKa = 4.1GNKK322 pKa = 10.26SITGSITGNTDD333 pKa = 3.13RR334 pKa = 11.84LVAAVNASGDD344 pKa = 4.06GVVSAIDD351 pKa = 3.62GLADD355 pKa = 3.9KK356 pKa = 11.15LKK358 pKa = 11.0LCDD361 pKa = 4.36PNTDD365 pKa = 3.45PFNCEE370 pKa = 3.87GEE372 pKa = 4.26NGLTPSSVEE381 pKa = 4.33SILKK385 pKa = 7.6QTSAVVTTSQVDD397 pKa = 3.44AEE399 pKa = 4.48EE400 pKa = 4.65GLLTTLKK407 pKa = 9.68EE408 pKa = 4.42TIDD411 pKa = 3.49NNLIEE416 pKa = 4.77DD417 pKa = 4.08TQSYY421 pKa = 9.8LEE423 pKa = 4.62DD424 pKa = 3.65MKK426 pKa = 11.49SDD428 pKa = 5.18LIGALPNSSQCDD440 pKa = 3.23VDD442 pKa = 4.21VLKK445 pKa = 10.08TPYY448 pKa = 10.58GDD450 pKa = 4.07FSIGCEE456 pKa = 3.59YY457 pKa = 10.67SARR460 pKa = 11.84LKK462 pKa = 10.96SILAFVFYY470 pKa = 10.25IYY472 pKa = 10.36TLYY475 pKa = 10.4TLAEE479 pKa = 3.96ILFTGVTPVAGTVPYY494 pKa = 10.16FSRR497 pKa = 11.84RR498 pKa = 3.44
MM1 pKa = 7.24RR2 pKa = 11.84TVLTLFSTTLLLSISFSSFAYY23 pKa = 9.53YY24 pKa = 10.02QLPFWPDD31 pKa = 2.7KK32 pKa = 11.01TYY34 pKa = 9.5LTPEE38 pKa = 4.01AAAAAYY44 pKa = 10.57LDD46 pKa = 3.57ILGGGSCKK54 pKa = 9.81RR55 pKa = 11.84DD56 pKa = 2.96TRR58 pKa = 11.84NFIKK62 pKa = 10.75ASISSINTPHH72 pKa = 6.43IAYY75 pKa = 10.31RR76 pKa = 11.84IDD78 pKa = 4.32KK79 pKa = 10.47YY80 pKa = 11.39LDD82 pKa = 3.65DD83 pKa = 3.77QCIVLSSTGNASVTLNVVDD102 pKa = 4.11NCPDD106 pKa = 3.18GTSPDD111 pKa = 4.62LSTGMCKK118 pKa = 10.24PKK120 pKa = 10.12PDD122 pKa = 3.45TPQYY126 pKa = 10.28CGTSAMFEE134 pKa = 4.36DD135 pKa = 4.93VANLYY140 pKa = 7.91NACYY144 pKa = 8.22EE145 pKa = 4.14QNGILSYY152 pKa = 10.61TCDD155 pKa = 3.69EE156 pKa = 4.29STQALDD162 pKa = 3.92AKK164 pKa = 10.71CDD166 pKa = 3.69LTTSDD171 pKa = 3.0QCVIGRR177 pKa = 11.84PTWPDD182 pKa = 3.63CLDD185 pKa = 4.38KK186 pKa = 10.83PHH188 pKa = 6.6QPNDD192 pKa = 3.62PTNPLPPVGGFNPSPVNPSVPPSPVEE218 pKa = 3.9KK219 pKa = 10.15PDD221 pKa = 3.28VQEE224 pKa = 4.03PDD226 pKa = 3.1KK227 pKa = 11.5TEE229 pKa = 3.84TSDD232 pKa = 3.31TGVINAIKK240 pKa = 10.52NLNDD244 pKa = 4.59DD245 pKa = 4.33LNKK248 pKa = 10.79SNTDD252 pKa = 2.65IHH254 pKa = 8.08NDD256 pKa = 3.02MNNIFSTMNDD266 pKa = 3.02ALRR269 pKa = 11.84QLNSTNTAIGQSIVEE284 pKa = 4.13QMKK287 pKa = 10.24QDD289 pKa = 3.35AQIYY293 pKa = 9.46DD294 pKa = 3.74NKK296 pKa = 10.73KK297 pKa = 9.53LQQQIAANNINAINSQTKK315 pKa = 10.42SLLEE319 pKa = 4.1GNKK322 pKa = 10.26SITGSITGNTDD333 pKa = 3.13RR334 pKa = 11.84LVAAVNASGDD344 pKa = 4.06GVVSAIDD351 pKa = 3.62GLADD355 pKa = 3.9KK356 pKa = 11.15LKK358 pKa = 11.0LCDD361 pKa = 4.36PNTDD365 pKa = 3.45PFNCEE370 pKa = 3.87GEE372 pKa = 4.26NGLTPSSVEE381 pKa = 4.33SILKK385 pKa = 7.6QTSAVVTTSQVDD397 pKa = 3.44AEE399 pKa = 4.48EE400 pKa = 4.65GLLTTLKK407 pKa = 9.68EE408 pKa = 4.42TIDD411 pKa = 3.49NNLIEE416 pKa = 4.77DD417 pKa = 4.08TQSYY421 pKa = 9.8LEE423 pKa = 4.62DD424 pKa = 3.65MKK426 pKa = 11.49SDD428 pKa = 5.18LIGALPNSSQCDD440 pKa = 3.23VDD442 pKa = 4.21VLKK445 pKa = 10.08TPYY448 pKa = 10.58GDD450 pKa = 4.07FSIGCEE456 pKa = 3.59YY457 pKa = 10.67SARR460 pKa = 11.84LKK462 pKa = 10.96SILAFVFYY470 pKa = 10.25IYY472 pKa = 10.36TLYY475 pKa = 10.4TLAEE479 pKa = 3.96ILFTGVTPVAGTVPYY494 pKa = 10.16FSRR497 pKa = 11.84RR498 pKa = 3.44
Molecular weight: 53.94 kDa
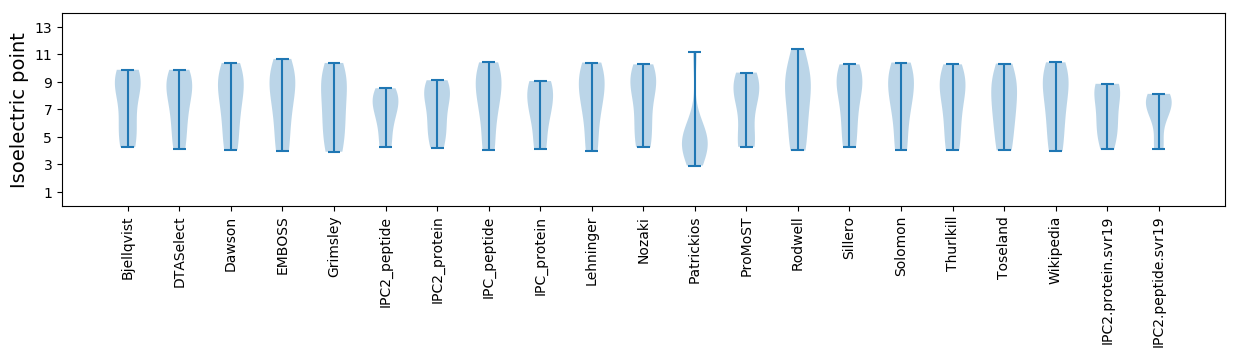
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|O80261|O80261_9VIRU Orf 498 OS=Vibrio phage fs2 OX=83201 PE=4 SV=1
MM1 pKa = 7.41KK2 pKa = 10.33KK3 pKa = 8.43STKK6 pKa = 10.04AALIVASLAAVAATGANAAIPQEE29 pKa = 4.14AQAALDD35 pKa = 3.65AVGEE39 pKa = 4.33FTSTVVGWMWGVGTAMIVGFVGLKK63 pKa = 9.2VVKK66 pKa = 10.45KK67 pKa = 9.96GANKK71 pKa = 8.84ATT73 pKa = 3.64
MM1 pKa = 7.41KK2 pKa = 10.33KK3 pKa = 8.43STKK6 pKa = 10.04AALIVASLAAVAATGANAAIPQEE29 pKa = 4.14AQAALDD35 pKa = 3.65AVGEE39 pKa = 4.33FTSTVVGWMWGVGTAMIVGFVGLKK63 pKa = 9.2VVKK66 pKa = 10.45KK67 pKa = 9.96GANKK71 pKa = 8.84ATT73 pKa = 3.64
Molecular weight: 7.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2445 |
38 |
716 |
271.7 |
30.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.344 ± 1.502 | 1.472 ± 0.363 |
5.644 ± 0.797 | 5.031 ± 0.853 |
4.663 ± 0.576 | 5.808 ± 0.496 |
1.554 ± 0.616 | 6.748 ± 0.679 |
5.89 ± 0.571 | 9.243 ± 0.688 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.209 ± 0.484 | 4.499 ± 0.712 |
4.54 ± 0.515 | 3.681 ± 0.342 |
4.663 ± 1.152 | 7.894 ± 0.816 |
6.83 ± 0.736 | 7.117 ± 0.904 |
1.35 ± 0.465 | 2.822 ± 0.403 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
