
Enterococcus phage vB_EfaP_IME199
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Rountreeviridae; Sarlesvirinae; Minhovirus; Enterococcus virus IME199
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
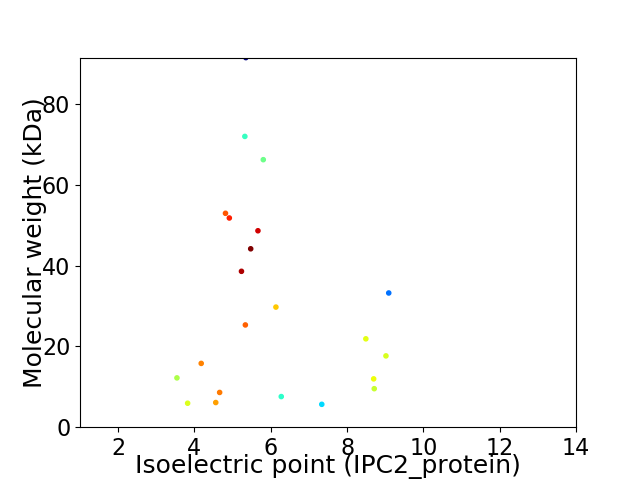
Virtual 2D-PAGE plot for 22 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0S2MYH8|A0A0S2MYH8_9CAUD DNA-directed DNA polymerase OS=Enterococcus phage vB_EfaP_IME199 OX=1747351 PE=4 SV=1
MM1 pKa = 7.59KK2 pKa = 10.23NFKK5 pKa = 8.37ITYY8 pKa = 7.31YY9 pKa = 10.52DD10 pKa = 3.0AWEE13 pKa = 4.46GQTMEE18 pKa = 4.58TYY20 pKa = 10.51IFAQSALEE28 pKa = 3.96AEE30 pKa = 4.82VIFTQEE36 pKa = 3.74EE37 pKa = 4.36DD38 pKa = 3.87SFCQVIDD45 pKa = 3.68VQEE48 pKa = 4.36EE49 pKa = 4.07II50 pKa = 4.53
MM1 pKa = 7.59KK2 pKa = 10.23NFKK5 pKa = 8.37ITYY8 pKa = 7.31YY9 pKa = 10.52DD10 pKa = 3.0AWEE13 pKa = 4.46GQTMEE18 pKa = 4.58TYY20 pKa = 10.51IFAQSALEE28 pKa = 3.96AEE30 pKa = 4.82VIFTQEE36 pKa = 3.74EE37 pKa = 4.36DD38 pKa = 3.87SFCQVIDD45 pKa = 3.68VQEE48 pKa = 4.36EE49 pKa = 4.07II50 pKa = 4.53
Molecular weight: 5.93 kDa
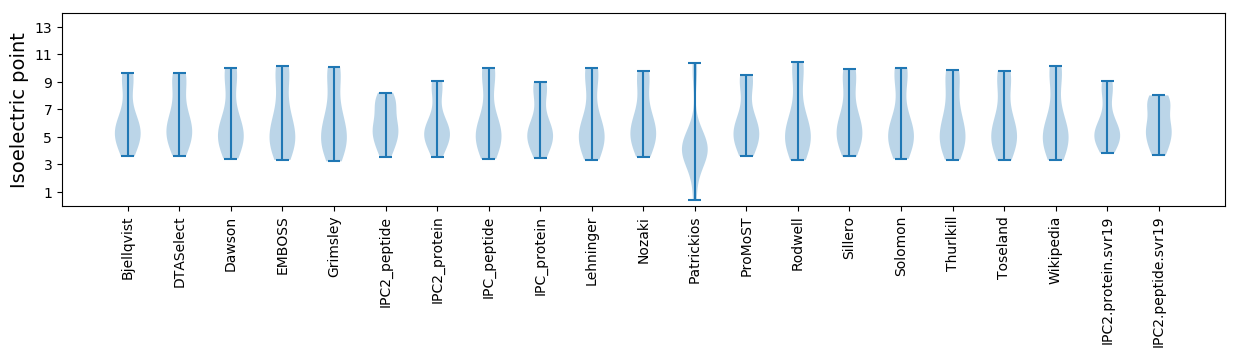
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0S2MYH6|A0A0S2MYH6_9CAUD Uncharacterized protein OS=Enterococcus phage vB_EfaP_IME199 OX=1747351 PE=4 SV=1
MM1 pKa = 6.48ITKK4 pKa = 10.14GIAGRR9 pKa = 11.84RR10 pKa = 11.84QGKK13 pKa = 8.92VKK15 pKa = 10.49GVVIHH20 pKa = 6.88NDD22 pKa = 3.06AGSNGATCSFYY33 pKa = 10.65RR34 pKa = 11.84GYY36 pKa = 9.44LTDD39 pKa = 4.5ANAAYY44 pKa = 9.86GFAHH48 pKa = 7.48WYY50 pKa = 8.24VASDD54 pKa = 4.71GILQVEE60 pKa = 4.99DD61 pKa = 3.98EE62 pKa = 5.46SNMAWHH68 pKa = 6.85TANSYY73 pKa = 11.23GNANFIGIEE82 pKa = 4.1ACQSMGNLAQFLEE95 pKa = 4.16NEE97 pKa = 3.67KK98 pKa = 10.65RR99 pKa = 11.84AIKK102 pKa = 10.09LAAEE106 pKa = 3.69ILKK109 pKa = 10.52RR110 pKa = 11.84HH111 pKa = 5.78GLTPNTTTVRR121 pKa = 11.84LHH123 pKa = 5.57NTFSSTACPHH133 pKa = 7.01RR134 pKa = 11.84SQEE137 pKa = 3.87QHH139 pKa = 5.55GAGSKK144 pKa = 7.22TQAYY148 pKa = 8.86FIAEE152 pKa = 4.37IKK154 pKa = 10.51KK155 pKa = 9.86HH156 pKa = 5.49MNGSTPAPSKK166 pKa = 9.53PTTPSQHH173 pKa = 6.68DD174 pKa = 3.84KK175 pKa = 11.13AVSASPAKK183 pKa = 10.3HH184 pKa = 6.06QGNAWGKK191 pKa = 7.85LDD193 pKa = 4.24KK194 pKa = 11.04YY195 pKa = 10.67RR196 pKa = 11.84EE197 pKa = 4.01EE198 pKa = 3.99PAGKK202 pKa = 9.59IRR204 pKa = 11.84VAGWLVPDD212 pKa = 4.95KK213 pKa = 10.62PQGPIGKK220 pKa = 8.29YY221 pKa = 9.59AYY223 pKa = 10.33VIFMQHH229 pKa = 5.07GTNKK233 pKa = 10.09EE234 pKa = 3.93LTRR237 pKa = 11.84VQSAGIKK244 pKa = 10.33RR245 pKa = 11.84PDD247 pKa = 3.14VKK249 pKa = 10.48KK250 pKa = 10.45AYY252 pKa = 9.82SYY254 pKa = 11.15QGGQEE259 pKa = 4.09LGFDD263 pKa = 3.76VTVKK267 pKa = 10.47KK268 pKa = 10.75AQFKK272 pKa = 9.54GKK274 pKa = 9.98KK275 pKa = 8.93VDD277 pKa = 3.93VILRR281 pKa = 11.84RR282 pKa = 11.84ANKK285 pKa = 10.27ANGEE289 pKa = 4.25VAVNDD294 pKa = 3.37VRR296 pKa = 11.84IDD298 pKa = 3.88SIYY301 pKa = 10.17LTLL304 pKa = 4.81
MM1 pKa = 6.48ITKK4 pKa = 10.14GIAGRR9 pKa = 11.84RR10 pKa = 11.84QGKK13 pKa = 8.92VKK15 pKa = 10.49GVVIHH20 pKa = 6.88NDD22 pKa = 3.06AGSNGATCSFYY33 pKa = 10.65RR34 pKa = 11.84GYY36 pKa = 9.44LTDD39 pKa = 4.5ANAAYY44 pKa = 9.86GFAHH48 pKa = 7.48WYY50 pKa = 8.24VASDD54 pKa = 4.71GILQVEE60 pKa = 4.99DD61 pKa = 3.98EE62 pKa = 5.46SNMAWHH68 pKa = 6.85TANSYY73 pKa = 11.23GNANFIGIEE82 pKa = 4.1ACQSMGNLAQFLEE95 pKa = 4.16NEE97 pKa = 3.67KK98 pKa = 10.65RR99 pKa = 11.84AIKK102 pKa = 10.09LAAEE106 pKa = 3.69ILKK109 pKa = 10.52RR110 pKa = 11.84HH111 pKa = 5.78GLTPNTTTVRR121 pKa = 11.84LHH123 pKa = 5.57NTFSSTACPHH133 pKa = 7.01RR134 pKa = 11.84SQEE137 pKa = 3.87QHH139 pKa = 5.55GAGSKK144 pKa = 7.22TQAYY148 pKa = 8.86FIAEE152 pKa = 4.37IKK154 pKa = 10.51KK155 pKa = 9.86HH156 pKa = 5.49MNGSTPAPSKK166 pKa = 9.53PTTPSQHH173 pKa = 6.68DD174 pKa = 3.84KK175 pKa = 11.13AVSASPAKK183 pKa = 10.3HH184 pKa = 6.06QGNAWGKK191 pKa = 7.85LDD193 pKa = 4.24KK194 pKa = 11.04YY195 pKa = 10.67RR196 pKa = 11.84EE197 pKa = 4.01EE198 pKa = 3.99PAGKK202 pKa = 9.59IRR204 pKa = 11.84VAGWLVPDD212 pKa = 4.95KK213 pKa = 10.62PQGPIGKK220 pKa = 8.29YY221 pKa = 9.59AYY223 pKa = 10.33VIFMQHH229 pKa = 5.07GTNKK233 pKa = 10.09EE234 pKa = 3.93LTRR237 pKa = 11.84VQSAGIKK244 pKa = 10.33RR245 pKa = 11.84PDD247 pKa = 3.14VKK249 pKa = 10.48KK250 pKa = 10.45AYY252 pKa = 9.82SYY254 pKa = 11.15QGGQEE259 pKa = 4.09LGFDD263 pKa = 3.76VTVKK267 pKa = 10.47KK268 pKa = 10.75AQFKK272 pKa = 9.54GKK274 pKa = 9.98KK275 pKa = 8.93VDD277 pKa = 3.93VILRR281 pKa = 11.84RR282 pKa = 11.84ANKK285 pKa = 10.27ANGEE289 pKa = 4.25VAVNDD294 pKa = 3.37VRR296 pKa = 11.84IDD298 pKa = 3.88SIYY301 pKa = 10.17LTLL304 pKa = 4.81
Molecular weight: 33.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5950 |
45 |
779 |
270.5 |
30.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.218 ± 0.537 | 0.655 ± 0.171 |
5.916 ± 0.203 | 7.059 ± 0.588 |
4.639 ± 0.402 | 6.017 ± 0.573 |
1.832 ± 0.25 | 6.437 ± 0.254 |
7.412 ± 0.451 | 7.832 ± 0.455 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.639 ± 0.153 | 7.496 ± 0.37 |
3.496 ± 0.363 | 4.067 ± 0.334 |
3.445 ± 0.315 | 5.798 ± 0.314 |
7.244 ± 0.474 | 5.697 ± 0.309 |
1.176 ± 0.13 | 4.924 ± 0.343 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
