
Dokdonia sinensis
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Dokdonia
Average proteome isoelectric point is 6.27
Get precalculated fractions of proteins
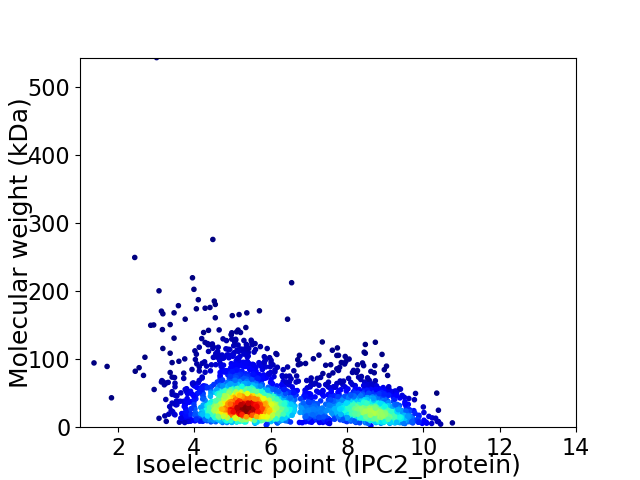
Virtual 2D-PAGE plot for 3191 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
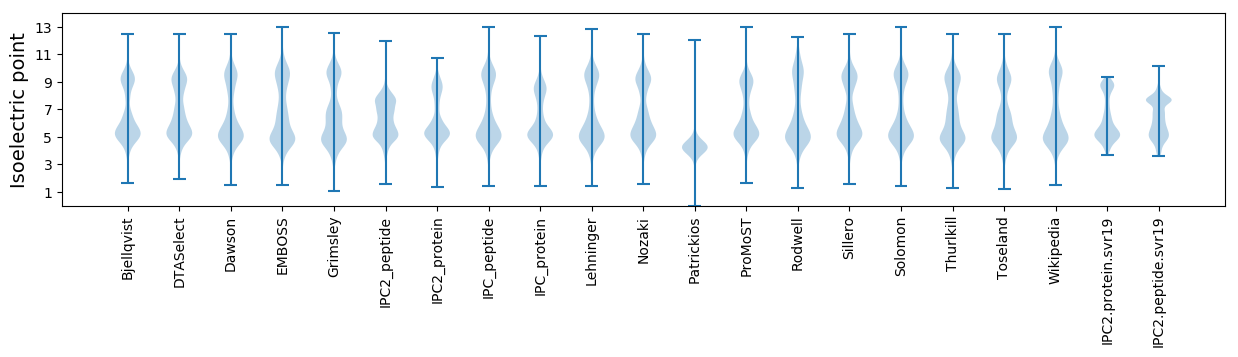
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3M0G828|A0A3M0G828_9FLAO Uncharacterized protein OS=Dokdonia sinensis OX=2479847 GN=EAX61_06975 PE=4 SV=1
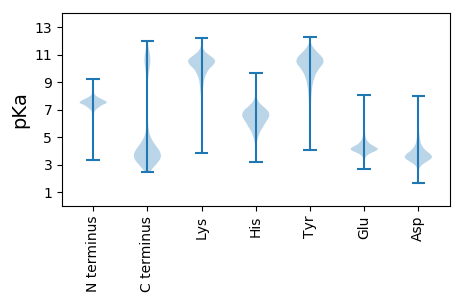
MM1 pKa = 7.5KK2 pKa = 10.15KK3 pKa = 10.04YY4 pKa = 10.41IKK6 pKa = 9.77YY7 pKa = 10.57LPVLAIAFAACEE19 pKa = 4.17PEE21 pKa = 3.65FDD23 pKa = 4.97NPIEE27 pKa = 4.09EE28 pKa = 5.41DD29 pKa = 3.5NFFTSGEE36 pKa = 4.13ADD38 pKa = 3.72FSNYY42 pKa = 8.17VALGNSLTAGFADD55 pKa = 4.0GALYY59 pKa = 8.33RR60 pKa = 11.84TAQNDD65 pKa = 3.88SYY67 pKa = 10.46PNIMAQQFALAGGGEE82 pKa = 4.35FTQPLTNDD90 pKa = 3.15NLGGLTLGGNQITANRR106 pKa = 11.84FVLATDD112 pKa = 3.85ANGNPGPAVLAGTPTTDD129 pKa = 2.59ISNVLSGPFNNMGVPGAKK147 pKa = 9.49SFHH150 pKa = 6.51LVTPGYY156 pKa = 10.89GNVANFPAAANPYY169 pKa = 8.83FIRR172 pKa = 11.84IASEE176 pKa = 3.72PNATVLADD184 pKa = 4.18AVAQNPSFFSLWIGNNDD201 pKa = 3.23VLSYY205 pKa = 9.37ATSGGAGTDD214 pKa = 3.14QTGNLDD220 pKa = 3.34PSTYY224 pKa = 10.36GGNDD228 pKa = 2.58ITDD231 pKa = 4.0PNVFASVYY239 pKa = 8.05SQQVAALVGTGAKK252 pKa = 9.73GALVNIADD260 pKa = 3.83VTSIPFFTTVPAQSIPLDD278 pKa = 3.81APTAAFLNSNFALYY292 pKa = 9.37NQQVLPGLVQFGIITAEE309 pKa = 3.74EE310 pKa = 3.6AAARR314 pKa = 11.84QINFAEE320 pKa = 4.73GVNFPTMSDD329 pKa = 3.88DD330 pKa = 4.24DD331 pKa = 4.36LTDD334 pKa = 3.39ISQILQGPPFGLDD347 pKa = 2.72AATAATLGTLRR358 pKa = 11.84QANDD362 pKa = 3.11SDD364 pKa = 5.05LFPLTAAGVLGTLVNDD380 pKa = 3.58NPQLINGVSVPLADD394 pKa = 3.64QFVLTAVEE402 pKa = 4.05QGRR405 pKa = 11.84VATAQASYY413 pKa = 10.87NATIQGLAAANDD425 pKa = 4.03LAYY428 pKa = 10.88VDD430 pKa = 5.78ARR432 pKa = 11.84AQLQQVAGSGVVFDD446 pKa = 5.07GGTLTSTFVTGGAFSLDD463 pKa = 3.32GVHH466 pKa = 6.96PSPRR470 pKa = 11.84GYY472 pKa = 11.29ALTANSIIDD481 pKa = 4.82AINAKK486 pKa = 10.32YY487 pKa = 10.03GATVPKK493 pKa = 10.67VNIGDD498 pKa = 3.77YY499 pKa = 9.74STVNITNNVNN509 pKa = 2.98
MM1 pKa = 7.5KK2 pKa = 10.15KK3 pKa = 10.04YY4 pKa = 10.41IKK6 pKa = 9.77YY7 pKa = 10.57LPVLAIAFAACEE19 pKa = 4.17PEE21 pKa = 3.65FDD23 pKa = 4.97NPIEE27 pKa = 4.09EE28 pKa = 5.41DD29 pKa = 3.5NFFTSGEE36 pKa = 4.13ADD38 pKa = 3.72FSNYY42 pKa = 8.17VALGNSLTAGFADD55 pKa = 4.0GALYY59 pKa = 8.33RR60 pKa = 11.84TAQNDD65 pKa = 3.88SYY67 pKa = 10.46PNIMAQQFALAGGGEE82 pKa = 4.35FTQPLTNDD90 pKa = 3.15NLGGLTLGGNQITANRR106 pKa = 11.84FVLATDD112 pKa = 3.85ANGNPGPAVLAGTPTTDD129 pKa = 2.59ISNVLSGPFNNMGVPGAKK147 pKa = 9.49SFHH150 pKa = 6.51LVTPGYY156 pKa = 10.89GNVANFPAAANPYY169 pKa = 8.83FIRR172 pKa = 11.84IASEE176 pKa = 3.72PNATVLADD184 pKa = 4.18AVAQNPSFFSLWIGNNDD201 pKa = 3.23VLSYY205 pKa = 9.37ATSGGAGTDD214 pKa = 3.14QTGNLDD220 pKa = 3.34PSTYY224 pKa = 10.36GGNDD228 pKa = 2.58ITDD231 pKa = 4.0PNVFASVYY239 pKa = 8.05SQQVAALVGTGAKK252 pKa = 9.73GALVNIADD260 pKa = 3.83VTSIPFFTTVPAQSIPLDD278 pKa = 3.81APTAAFLNSNFALYY292 pKa = 9.37NQQVLPGLVQFGIITAEE309 pKa = 3.74EE310 pKa = 3.6AAARR314 pKa = 11.84QINFAEE320 pKa = 4.73GVNFPTMSDD329 pKa = 3.88DD330 pKa = 4.24DD331 pKa = 4.36LTDD334 pKa = 3.39ISQILQGPPFGLDD347 pKa = 2.72AATAATLGTLRR358 pKa = 11.84QANDD362 pKa = 3.11SDD364 pKa = 5.05LFPLTAAGVLGTLVNDD380 pKa = 3.58NPQLINGVSVPLADD394 pKa = 3.64QFVLTAVEE402 pKa = 4.05QGRR405 pKa = 11.84VATAQASYY413 pKa = 10.87NATIQGLAAANDD425 pKa = 4.03LAYY428 pKa = 10.88VDD430 pKa = 5.78ARR432 pKa = 11.84AQLQQVAGSGVVFDD446 pKa = 5.07GGTLTSTFVTGGAFSLDD463 pKa = 3.32GVHH466 pKa = 6.96PSPRR470 pKa = 11.84GYY472 pKa = 11.29ALTANSIIDD481 pKa = 4.82AINAKK486 pKa = 10.32YY487 pKa = 10.03GATVPKK493 pKa = 10.67VNIGDD498 pKa = 3.77YY499 pKa = 9.74STVNITNNVNN509 pKa = 2.98
Molecular weight: 52.56 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3M0G7T5|A0A3M0G7T5_9FLAO Eco47II family restriction endonuclease OS=Dokdonia sinensis OX=2479847 GN=EAX61_05690 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.92GRR40 pKa = 11.84KK41 pKa = 8.34KK42 pKa = 10.22ISVSSEE48 pKa = 3.61LRR50 pKa = 11.84HH51 pKa = 6.16KK52 pKa = 10.53KK53 pKa = 10.04
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.92GRR40 pKa = 11.84KK41 pKa = 8.34KK42 pKa = 10.22ISVSSEE48 pKa = 3.61LRR50 pKa = 11.84HH51 pKa = 6.16KK52 pKa = 10.53KK53 pKa = 10.04
Molecular weight: 6.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1089628 |
18 |
5232 |
341.5 |
38.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.105 ± 0.039 | 0.724 ± 0.016 |
6.197 ± 0.064 | 6.59 ± 0.039 |
5.07 ± 0.034 | 6.801 ± 0.042 |
1.717 ± 0.022 | 7.377 ± 0.035 |
6.739 ± 0.067 | 9.085 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.251 ± 0.022 | 5.562 ± 0.046 |
3.545 ± 0.023 | 3.489 ± 0.028 |
3.893 ± 0.037 | 6.176 ± 0.032 |
6.259 ± 0.065 | 6.465 ± 0.034 |
1.042 ± 0.015 | 3.912 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
