
Nocardioides sp. GY 10113
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Propionibacteriales; Nocardioidaceae; Nocardioides; unclassified Nocardioides
Average proteome isoelectric point is 5.91
Get precalculated fractions of proteins
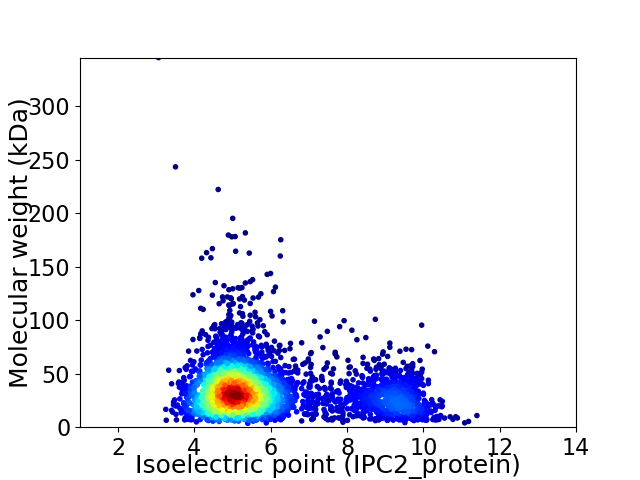
Virtual 2D-PAGE plot for 3866 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4T0V9S9|A0A4T0V9S9_9ACTN Indole-3-glycerol phosphate synthase OS=Nocardioides sp. GY 10113 OX=2569761 GN=trpC PE=3 SV=1
MM1 pKa = 8.0RR2 pKa = 11.84ARR4 pKa = 11.84NKK6 pKa = 9.61FAAVLGGTVVMASAAIVTTTTAASAVEE33 pKa = 4.07PTGGCWVYY41 pKa = 10.07NTGGNTNLEE50 pKa = 4.3SAPTSSISSALAGWSSSPPDD70 pKa = 3.52YY71 pKa = 11.44DD72 pKa = 3.51LTTSGGTLVGEE83 pKa = 4.29TRR85 pKa = 11.84NFSLVVSAGPSSPVPATTTAYY106 pKa = 10.89YY107 pKa = 9.95YY108 pKa = 10.72FSVNGANLAPVAVADD123 pKa = 4.59IPASTNTPGGTVTGSFDD140 pKa = 3.08IAQGGTNTLVLRR152 pKa = 11.84KK153 pKa = 9.69VIYY156 pKa = 9.62DD157 pKa = 3.29IPSYY161 pKa = 9.49SVRR164 pKa = 11.84VQCNGQSSGSQTGINPATSPVDD186 pKa = 3.73TNVTASFTAFAQGVATITGITNQTVMNAARR216 pKa = 11.84AGDD219 pKa = 4.17TISFAASNFSGADD232 pKa = 3.26ANSTAEE238 pKa = 3.62ICTTDD243 pKa = 3.03GATCVGATSFGVGTNGAGTGTITVPSGVSGSMRR276 pKa = 11.84LRR278 pKa = 11.84LTSGGEE284 pKa = 3.58VGMRR288 pKa = 11.84SITILGQPTLTLSLTGGGAGTTVGVTGTDD317 pKa = 3.09WDD319 pKa = 4.15PSQTVSIGGYY329 pKa = 5.36TAPLGGFPLPSTTDD343 pKa = 3.55PVSTATASATGTLSGSFTVNDD364 pKa = 3.97ANTAYY369 pKa = 9.68ISASQRR375 pKa = 11.84SGPGAIWSDD384 pKa = 2.95AAFAFSADD392 pKa = 2.96TCTAKK397 pKa = 10.65EE398 pKa = 3.95GLEE401 pKa = 4.38DD402 pKa = 4.19NSDD405 pKa = 3.46ACSVLEE411 pKa = 4.33TIEE414 pKa = 4.35LTVLPGDD421 pKa = 3.54LTMAKK426 pKa = 10.24AAGTVEE432 pKa = 3.99MGAVQLNGTDD442 pKa = 3.54QTSTGSLQDD451 pKa = 3.53VTVMDD456 pKa = 4.07YY457 pKa = 11.12RR458 pKa = 11.84GGTLGWSLIGTFAGLTSGPQPIDD481 pKa = 3.28ADD483 pKa = 3.92KK484 pKa = 11.37LSWTPSCTPGANSDD498 pKa = 3.68DD499 pKa = 4.0TVTVGAAGAFPSATEE514 pKa = 4.06ALPLCAVAAAGFGPDD529 pKa = 3.67GVSGGDD535 pKa = 3.27VTADD539 pKa = 3.31ADD541 pKa = 4.22LSLDD545 pKa = 4.39LLINQQAGDD554 pKa = 3.75YY555 pKa = 10.68SGTLTLTLSS564 pKa = 3.51
MM1 pKa = 8.0RR2 pKa = 11.84ARR4 pKa = 11.84NKK6 pKa = 9.61FAAVLGGTVVMASAAIVTTTTAASAVEE33 pKa = 4.07PTGGCWVYY41 pKa = 10.07NTGGNTNLEE50 pKa = 4.3SAPTSSISSALAGWSSSPPDD70 pKa = 3.52YY71 pKa = 11.44DD72 pKa = 3.51LTTSGGTLVGEE83 pKa = 4.29TRR85 pKa = 11.84NFSLVVSAGPSSPVPATTTAYY106 pKa = 10.89YY107 pKa = 9.95YY108 pKa = 10.72FSVNGANLAPVAVADD123 pKa = 4.59IPASTNTPGGTVTGSFDD140 pKa = 3.08IAQGGTNTLVLRR152 pKa = 11.84KK153 pKa = 9.69VIYY156 pKa = 9.62DD157 pKa = 3.29IPSYY161 pKa = 9.49SVRR164 pKa = 11.84VQCNGQSSGSQTGINPATSPVDD186 pKa = 3.73TNVTASFTAFAQGVATITGITNQTVMNAARR216 pKa = 11.84AGDD219 pKa = 4.17TISFAASNFSGADD232 pKa = 3.26ANSTAEE238 pKa = 3.62ICTTDD243 pKa = 3.03GATCVGATSFGVGTNGAGTGTITVPSGVSGSMRR276 pKa = 11.84LRR278 pKa = 11.84LTSGGEE284 pKa = 3.58VGMRR288 pKa = 11.84SITILGQPTLTLSLTGGGAGTTVGVTGTDD317 pKa = 3.09WDD319 pKa = 4.15PSQTVSIGGYY329 pKa = 5.36TAPLGGFPLPSTTDD343 pKa = 3.55PVSTATASATGTLSGSFTVNDD364 pKa = 3.97ANTAYY369 pKa = 9.68ISASQRR375 pKa = 11.84SGPGAIWSDD384 pKa = 2.95AAFAFSADD392 pKa = 2.96TCTAKK397 pKa = 10.65EE398 pKa = 3.95GLEE401 pKa = 4.38DD402 pKa = 4.19NSDD405 pKa = 3.46ACSVLEE411 pKa = 4.33TIEE414 pKa = 4.35LTVLPGDD421 pKa = 3.54LTMAKK426 pKa = 10.24AAGTVEE432 pKa = 3.99MGAVQLNGTDD442 pKa = 3.54QTSTGSLQDD451 pKa = 3.53VTVMDD456 pKa = 4.07YY457 pKa = 11.12RR458 pKa = 11.84GGTLGWSLIGTFAGLTSGPQPIDD481 pKa = 3.28ADD483 pKa = 3.92KK484 pKa = 11.37LSWTPSCTPGANSDD498 pKa = 3.68DD499 pKa = 4.0TVTVGAAGAFPSATEE514 pKa = 4.06ALPLCAVAAAGFGPDD529 pKa = 3.67GVSGGDD535 pKa = 3.27VTADD539 pKa = 3.31ADD541 pKa = 4.22LSLDD545 pKa = 4.39LLINQQAGDD554 pKa = 3.75YY555 pKa = 10.68SGTLTLTLSS564 pKa = 3.51
Molecular weight: 55.71 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4T0V0P3|A0A4T0V0P3_9ACTN Alpha/beta hydrolase OS=Nocardioides sp. GY 10113 OX=2569761 GN=E8D34_13455 PE=4 SV=1
MM1 pKa = 7.56AGSSPRR7 pKa = 11.84WRR9 pKa = 11.84LSSRR13 pKa = 11.84SEE15 pKa = 4.33RR16 pKa = 11.84AWSWAPRR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84PSPRR29 pKa = 11.84RR30 pKa = 11.84PRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84PTPRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84SSPRR45 pKa = 11.84RR46 pKa = 11.84WPRR49 pKa = 11.84PRR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84PARR56 pKa = 11.84RR57 pKa = 11.84PSATPPGRR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84SATPPSGRR75 pKa = 11.84RR76 pKa = 11.84RR77 pKa = 11.84GPSSRR82 pKa = 11.84PAGRR86 pKa = 11.84PPAAPPTGG94 pKa = 3.38
MM1 pKa = 7.56AGSSPRR7 pKa = 11.84WRR9 pKa = 11.84LSSRR13 pKa = 11.84SEE15 pKa = 4.33RR16 pKa = 11.84AWSWAPRR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84PSPRR29 pKa = 11.84RR30 pKa = 11.84PRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84PTPRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84SSPRR45 pKa = 11.84RR46 pKa = 11.84WPRR49 pKa = 11.84PRR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84PARR56 pKa = 11.84RR57 pKa = 11.84PSATPPGRR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84SATPPSGRR75 pKa = 11.84RR76 pKa = 11.84RR77 pKa = 11.84GPSSRR82 pKa = 11.84PAGRR86 pKa = 11.84PPAAPPTGG94 pKa = 3.38
Molecular weight: 10.88 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1303206 |
33 |
3357 |
337.1 |
35.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.95 ± 0.064 | 0.705 ± 0.01 |
6.413 ± 0.035 | 5.84 ± 0.041 |
2.746 ± 0.02 | 9.541 ± 0.036 |
2.08 ± 0.02 | 3.45 ± 0.026 |
1.789 ± 0.028 | 10.17 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.762 ± 0.015 | 1.701 ± 0.022 |
5.766 ± 0.031 | 2.555 ± 0.02 |
7.727 ± 0.044 | 4.995 ± 0.029 |
6.018 ± 0.038 | 9.28 ± 0.036 |
1.516 ± 0.017 | 1.997 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
