
Setaria italica (Foxtail millet) (Panicum italicum)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae;
Average proteome isoelectric point is 6.94
Get precalculated fractions of proteins
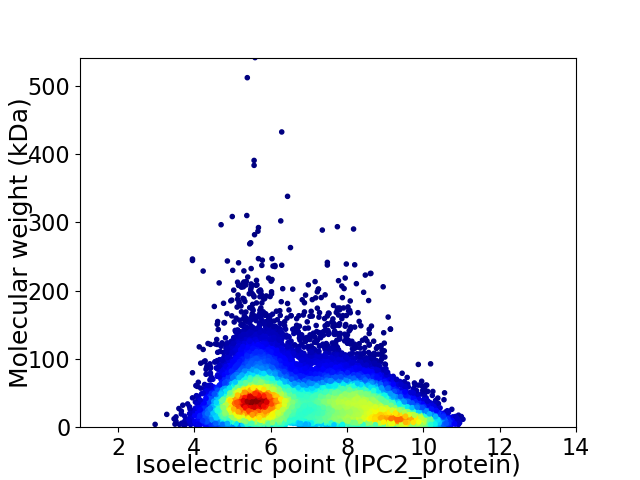
Virtual 2D-PAGE plot for 39467 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K3XFV8|K3XFV8_SETIT Aspartate--tRNA ligase OS=Setaria italica OX=4555 PE=3 SV=1
MM1 pKa = 7.81PDD3 pKa = 2.6WRR5 pKa = 11.84VGEE8 pKa = 4.22FEE10 pKa = 5.44GKK12 pKa = 8.49FTDD15 pKa = 5.31EE16 pKa = 4.27FAQSNRR22 pKa = 11.84SEE24 pKa = 4.16HH25 pKa = 6.04EE26 pKa = 4.26SGIEE30 pKa = 3.92APDD33 pKa = 3.44VTSSKK38 pKa = 10.47KK39 pKa = 10.18LKK41 pKa = 10.12HH42 pKa = 5.74AAASEE47 pKa = 4.49KK48 pKa = 9.84IHH50 pKa = 6.27QGVISGTNDD59 pKa = 3.28SDD61 pKa = 3.97SQKK64 pKa = 10.72CNSEE68 pKa = 4.68HH69 pKa = 5.78IQCANGNINSNGDD82 pKa = 3.71CKK84 pKa = 10.95DD85 pKa = 3.06GSNAFTSRR93 pKa = 11.84EE94 pKa = 3.84EE95 pKa = 3.87NTIVEE100 pKa = 4.71TRR102 pKa = 11.84CPTDD106 pKa = 2.59NWNSCQFALSNGSSILNNHH125 pKa = 6.3SAPQDD130 pKa = 3.42SLAYY134 pKa = 10.25GDD136 pKa = 4.7NDD138 pKa = 4.25LNYY141 pKa = 9.66IDD143 pKa = 5.09WPGIDD148 pKa = 3.64NFEE151 pKa = 4.89DD152 pKa = 3.42VDD154 pKa = 4.03TLFRR158 pKa = 11.84RR159 pKa = 11.84SDD161 pKa = 3.22STYY164 pKa = 10.89GQQQLEE170 pKa = 4.4NTDD173 pKa = 3.54GLSWIPSSSDD183 pKa = 2.81AVYY186 pKa = 10.86SSDD189 pKa = 3.78VALQQGFGSSYY200 pKa = 10.8SDD202 pKa = 3.52YY203 pKa = 11.45GILDD207 pKa = 4.22DD208 pKa = 6.54LSAFQCAEE216 pKa = 4.21DD217 pKa = 3.98KK218 pKa = 10.95SLPSVDD224 pKa = 4.39PSAALCDD231 pKa = 3.64NQFDD235 pKa = 3.98DD236 pKa = 4.4TYY238 pKa = 10.95MFSEE242 pKa = 4.15QKK244 pKa = 10.46NVNGYY249 pKa = 10.31GDD251 pKa = 3.77QIYY254 pKa = 10.02QEE256 pKa = 5.07DD257 pKa = 4.42VMEE260 pKa = 5.44LLPTDD265 pKa = 4.08QICNGNGNIDD275 pKa = 3.42MM276 pKa = 5.44
MM1 pKa = 7.81PDD3 pKa = 2.6WRR5 pKa = 11.84VGEE8 pKa = 4.22FEE10 pKa = 5.44GKK12 pKa = 8.49FTDD15 pKa = 5.31EE16 pKa = 4.27FAQSNRR22 pKa = 11.84SEE24 pKa = 4.16HH25 pKa = 6.04EE26 pKa = 4.26SGIEE30 pKa = 3.92APDD33 pKa = 3.44VTSSKK38 pKa = 10.47KK39 pKa = 10.18LKK41 pKa = 10.12HH42 pKa = 5.74AAASEE47 pKa = 4.49KK48 pKa = 9.84IHH50 pKa = 6.27QGVISGTNDD59 pKa = 3.28SDD61 pKa = 3.97SQKK64 pKa = 10.72CNSEE68 pKa = 4.68HH69 pKa = 5.78IQCANGNINSNGDD82 pKa = 3.71CKK84 pKa = 10.95DD85 pKa = 3.06GSNAFTSRR93 pKa = 11.84EE94 pKa = 3.84EE95 pKa = 3.87NTIVEE100 pKa = 4.71TRR102 pKa = 11.84CPTDD106 pKa = 2.59NWNSCQFALSNGSSILNNHH125 pKa = 6.3SAPQDD130 pKa = 3.42SLAYY134 pKa = 10.25GDD136 pKa = 4.7NDD138 pKa = 4.25LNYY141 pKa = 9.66IDD143 pKa = 5.09WPGIDD148 pKa = 3.64NFEE151 pKa = 4.89DD152 pKa = 3.42VDD154 pKa = 4.03TLFRR158 pKa = 11.84RR159 pKa = 11.84SDD161 pKa = 3.22STYY164 pKa = 10.89GQQQLEE170 pKa = 4.4NTDD173 pKa = 3.54GLSWIPSSSDD183 pKa = 2.81AVYY186 pKa = 10.86SSDD189 pKa = 3.78VALQQGFGSSYY200 pKa = 10.8SDD202 pKa = 3.52YY203 pKa = 11.45GILDD207 pKa = 4.22DD208 pKa = 6.54LSAFQCAEE216 pKa = 4.21DD217 pKa = 3.98KK218 pKa = 10.95SLPSVDD224 pKa = 4.39PSAALCDD231 pKa = 3.64NQFDD235 pKa = 3.98DD236 pKa = 4.4TYY238 pKa = 10.95MFSEE242 pKa = 4.15QKK244 pKa = 10.46NVNGYY249 pKa = 10.31GDD251 pKa = 3.77QIYY254 pKa = 10.02QEE256 pKa = 5.07DD257 pKa = 4.42VMEE260 pKa = 5.44LLPTDD265 pKa = 4.08QICNGNGNIDD275 pKa = 3.42MM276 pKa = 5.44
Molecular weight: 30.34 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K3Y448|K3Y448_SETIT Uncharacterized protein OS=Setaria italica OX=4555 PE=4 SV=1
MM1 pKa = 7.33RR2 pKa = 11.84CRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84GKK13 pKa = 10.15CSGGRR18 pKa = 11.84RR19 pKa = 11.84GTRR22 pKa = 11.84ARR24 pKa = 11.84GQRR27 pKa = 11.84GTPPTGARR35 pKa = 11.84SRR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84GPRR43 pKa = 11.84LWDD46 pKa = 3.09ARR48 pKa = 11.84MGDD51 pKa = 3.43GTPRR55 pKa = 11.84TRR57 pKa = 11.84RR58 pKa = 11.84LPRR61 pKa = 11.84RR62 pKa = 11.84ARR64 pKa = 11.84RR65 pKa = 11.84GRR67 pKa = 11.84WRR69 pKa = 11.84NGGTSGRR76 pKa = 11.84TRR78 pKa = 11.84SPPPQSRR85 pKa = 11.84NRR87 pKa = 11.84HH88 pKa = 3.73GRR90 pKa = 11.84TRR92 pKa = 11.84RR93 pKa = 11.84GKK95 pKa = 10.57ASGPLL100 pKa = 3.39
MM1 pKa = 7.33RR2 pKa = 11.84CRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84GKK13 pKa = 10.15CSGGRR18 pKa = 11.84RR19 pKa = 11.84GTRR22 pKa = 11.84ARR24 pKa = 11.84GQRR27 pKa = 11.84GTPPTGARR35 pKa = 11.84SRR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84GPRR43 pKa = 11.84LWDD46 pKa = 3.09ARR48 pKa = 11.84MGDD51 pKa = 3.43GTPRR55 pKa = 11.84TRR57 pKa = 11.84RR58 pKa = 11.84LPRR61 pKa = 11.84RR62 pKa = 11.84ARR64 pKa = 11.84RR65 pKa = 11.84GRR67 pKa = 11.84WRR69 pKa = 11.84NGGTSGRR76 pKa = 11.84TRR78 pKa = 11.84SPPPQSRR85 pKa = 11.84NRR87 pKa = 11.84HH88 pKa = 3.73GRR90 pKa = 11.84TRR92 pKa = 11.84RR93 pKa = 11.84GKK95 pKa = 10.57ASGPLL100 pKa = 3.39
Molecular weight: 11.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
14674120 |
8 |
5181 |
371.8 |
40.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.269 ± 0.021 | 1.948 ± 0.006 |
5.367 ± 0.011 | 5.979 ± 0.014 |
3.718 ± 0.008 | 7.295 ± 0.013 |
2.494 ± 0.006 | 4.41 ± 0.01 |
4.918 ± 0.013 | 9.598 ± 0.017 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.422 ± 0.006 | 3.509 ± 0.009 |
5.65 ± 0.015 | 3.452 ± 0.008 |
6.323 ± 0.012 | 8.153 ± 0.015 |
4.762 ± 0.008 | 6.769 ± 0.01 |
1.331 ± 0.004 | 2.63 ± 0.007 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
