
Pseudogymnoascus sp. WSF 3629
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Leotiomycetes; Leotiomycetes incertae sedis; Pseudeurotiaceae; Pseudogymnoascus; unclassified Pseudogymnoascus
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
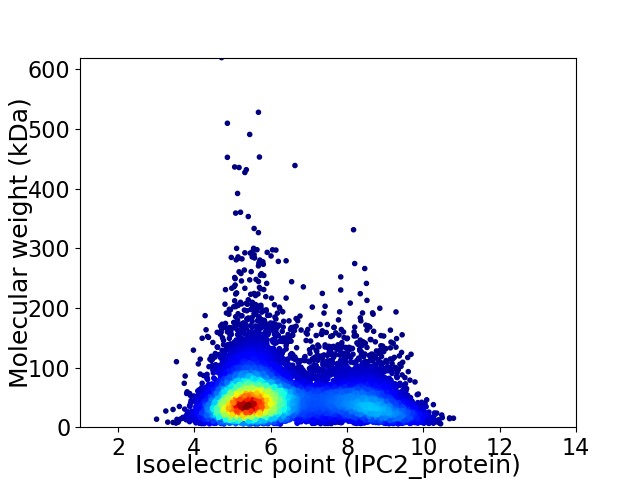
Virtual 2D-PAGE plot for 11000 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B8CQI2|A0A1B8CQI2_9PEZI Uncharacterized protein OS=Pseudogymnoascus sp. WSF 3629 OX=1622147 GN=VE00_00111 PE=4 SV=1
MM1 pKa = 7.89RR2 pKa = 11.84GLLFLTLAANLGSAFTQVVDD22 pKa = 4.5LGLAASYY29 pKa = 10.83GVLAHH34 pKa = 7.05ASISNSGPTTVNGDD48 pKa = 2.81IGTTGTSIVGFPPGVYY64 pKa = 9.06TGNRR68 pKa = 11.84NVGLLATTAFNNAEE82 pKa = 3.94AAYY85 pKa = 6.44TTLGQLPSIILVGNLGGRR103 pKa = 11.84LLRR106 pKa = 11.84PGVYY110 pKa = 9.67RR111 pKa = 11.84FSTSAVLSGTLILAGQGSPCDD132 pKa = 3.0SWVFLIGSTLITSVGSSVLVTGGGNPGNVFWRR164 pKa = 11.84VGSSATIQIGSQFSGNILAGVAVTLNSGASIQGSVYY200 pKa = 10.73ALGSSVVLNSNEE212 pKa = 3.56VAAQEE217 pKa = 4.12NSCPLGASTSTSVSSTSTTASSDD240 pKa = 3.19SSTSTDD246 pKa = 2.67VSTTTDD252 pKa = 2.87SSTSTTTDD260 pKa = 2.7VTTTTSSDD268 pKa = 3.09SSTSTDD274 pKa = 2.69VSTTSTSTDD283 pKa = 2.96VPTTTDD289 pKa = 2.79SSTSTDD295 pKa = 2.9VSTTTSTSTDD305 pKa = 3.07VPTTTDD311 pKa = 2.79SSTSTDD317 pKa = 3.11VTTTTSTSTDD327 pKa = 3.07VPTTTDD333 pKa = 2.79SSTSTDD339 pKa = 3.11VTTTTSTSTDD349 pKa = 2.83VSTTDD354 pKa = 2.9VSTTTDD360 pKa = 2.62ITTATSTTTDD370 pKa = 2.97VTTATSTSTDD380 pKa = 2.74VSTTTDD386 pKa = 2.56ITTATDD392 pKa = 2.97VTTATDD398 pKa = 2.92ITTATNITTAIDD410 pKa = 3.05ITTATDD416 pKa = 2.65ITTATDD422 pKa = 2.72ITTATDD428 pKa = 2.97VTTATDD434 pKa = 3.04ITTATSTSTDD444 pKa = 3.08VTTTTSTSTDD454 pKa = 3.07VPTTTDD460 pKa = 3.03SSSSSSSVTSASSSLTSTSMASLPSLTSSTIISVTNSSAVSSPPASTTSTVYY512 pKa = 9.67ATTVYY517 pKa = 10.51TITKK521 pKa = 10.05CPATVTNCPIGSTTTEE537 pKa = 4.72LISLYY542 pKa = 8.45TTVCPVAASATKK554 pKa = 10.05QPPSGYY560 pKa = 8.25TVSTVFTTIVYY571 pKa = 8.07TITKK575 pKa = 9.52CPKK578 pKa = 9.13TVTNCPVGSVTTEE591 pKa = 3.17IRR593 pKa = 11.84SLYY596 pKa = 7.48TTTCPVTQSFVNSVLPPQSSIPARR620 pKa = 11.84PASSQLPAAVLPSRR634 pKa = 11.84ISSIAIASPVSTVSPALGLSGAATVSSGAAPVSSVSPASGPSGVPGSVPLIGAPPSSNLVISTVTASKK702 pKa = 9.29GTANVSQTSQPSAAYY717 pKa = 7.41TGPIASSGVMNKK729 pKa = 8.04GTSASLLVLAIGALLFLL746 pKa = 5.21
MM1 pKa = 7.89RR2 pKa = 11.84GLLFLTLAANLGSAFTQVVDD22 pKa = 4.5LGLAASYY29 pKa = 10.83GVLAHH34 pKa = 7.05ASISNSGPTTVNGDD48 pKa = 2.81IGTTGTSIVGFPPGVYY64 pKa = 9.06TGNRR68 pKa = 11.84NVGLLATTAFNNAEE82 pKa = 3.94AAYY85 pKa = 6.44TTLGQLPSIILVGNLGGRR103 pKa = 11.84LLRR106 pKa = 11.84PGVYY110 pKa = 9.67RR111 pKa = 11.84FSTSAVLSGTLILAGQGSPCDD132 pKa = 3.0SWVFLIGSTLITSVGSSVLVTGGGNPGNVFWRR164 pKa = 11.84VGSSATIQIGSQFSGNILAGVAVTLNSGASIQGSVYY200 pKa = 10.73ALGSSVVLNSNEE212 pKa = 3.56VAAQEE217 pKa = 4.12NSCPLGASTSTSVSSTSTTASSDD240 pKa = 3.19SSTSTDD246 pKa = 2.67VSTTTDD252 pKa = 2.87SSTSTTTDD260 pKa = 2.7VTTTTSSDD268 pKa = 3.09SSTSTDD274 pKa = 2.69VSTTSTSTDD283 pKa = 2.96VPTTTDD289 pKa = 2.79SSTSTDD295 pKa = 2.9VSTTTSTSTDD305 pKa = 3.07VPTTTDD311 pKa = 2.79SSTSTDD317 pKa = 3.11VTTTTSTSTDD327 pKa = 3.07VPTTTDD333 pKa = 2.79SSTSTDD339 pKa = 3.11VTTTTSTSTDD349 pKa = 2.83VSTTDD354 pKa = 2.9VSTTTDD360 pKa = 2.62ITTATSTTTDD370 pKa = 2.97VTTATSTSTDD380 pKa = 2.74VSTTTDD386 pKa = 2.56ITTATDD392 pKa = 2.97VTTATDD398 pKa = 2.92ITTATNITTAIDD410 pKa = 3.05ITTATDD416 pKa = 2.65ITTATDD422 pKa = 2.72ITTATDD428 pKa = 2.97VTTATDD434 pKa = 3.04ITTATSTSTDD444 pKa = 3.08VTTTTSTSTDD454 pKa = 3.07VPTTTDD460 pKa = 3.03SSSSSSSVTSASSSLTSTSMASLPSLTSSTIISVTNSSAVSSPPASTTSTVYY512 pKa = 9.67ATTVYY517 pKa = 10.51TITKK521 pKa = 10.05CPATVTNCPIGSTTTEE537 pKa = 4.72LISLYY542 pKa = 8.45TTVCPVAASATKK554 pKa = 10.05QPPSGYY560 pKa = 8.25TVSTVFTTIVYY571 pKa = 8.07TITKK575 pKa = 9.52CPKK578 pKa = 9.13TVTNCPVGSVTTEE591 pKa = 3.17IRR593 pKa = 11.84SLYY596 pKa = 7.48TTTCPVTQSFVNSVLPPQSSIPARR620 pKa = 11.84PASSQLPAAVLPSRR634 pKa = 11.84ISSIAIASPVSTVSPALGLSGAATVSSGAAPVSSVSPASGPSGVPGSVPLIGAPPSSNLVISTVTASKK702 pKa = 9.29GTANVSQTSQPSAAYY717 pKa = 7.41TGPIASSGVMNKK729 pKa = 8.04GTSASLLVLAIGALLFLL746 pKa = 5.21
Molecular weight: 73.6 kDa
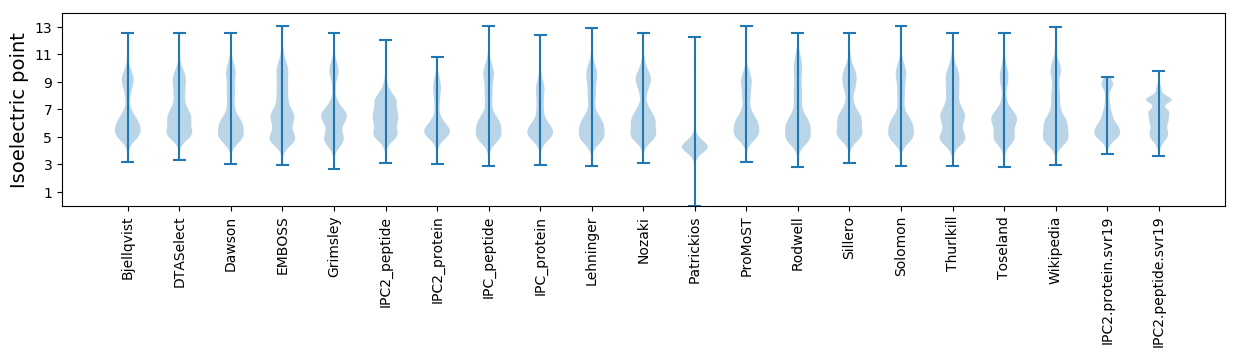
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B8CG56|A0A1B8CG56_9PEZI Glucanase OS=Pseudogymnoascus sp. WSF 3629 OX=1622147 GN=VE00_03807 PE=3 SV=1
MM1 pKa = 7.4FSLRR5 pKa = 11.84SSRR8 pKa = 11.84GVTAALKK15 pKa = 10.29PVMQARR21 pKa = 11.84AVKK24 pKa = 10.25SVAPQASRR32 pKa = 11.84TFSILTPLRR41 pKa = 11.84PSLTSSFAPRR51 pKa = 11.84ASAALDD57 pKa = 3.58TTTTTSTTTSAAGTTTILDD76 pKa = 4.59LLPKK80 pKa = 10.19ISTHH84 pKa = 6.55PSLAGIQVRR93 pKa = 11.84CGPRR97 pKa = 11.84NTFSPSHH104 pKa = 6.08FVRR107 pKa = 11.84KK108 pKa = 9.24RR109 pKa = 11.84RR110 pKa = 11.84HH111 pKa = 5.18GFLSRR116 pKa = 11.84VRR118 pKa = 11.84TRR120 pKa = 11.84KK121 pKa = 9.59GRR123 pKa = 11.84ATLQRR128 pKa = 11.84RR129 pKa = 11.84KK130 pKa = 10.21SKK132 pKa = 10.49NRR134 pKa = 11.84STLSHH139 pKa = 6.67
MM1 pKa = 7.4FSLRR5 pKa = 11.84SSRR8 pKa = 11.84GVTAALKK15 pKa = 10.29PVMQARR21 pKa = 11.84AVKK24 pKa = 10.25SVAPQASRR32 pKa = 11.84TFSILTPLRR41 pKa = 11.84PSLTSSFAPRR51 pKa = 11.84ASAALDD57 pKa = 3.58TTTTTSTTTSAAGTTTILDD76 pKa = 4.59LLPKK80 pKa = 10.19ISTHH84 pKa = 6.55PSLAGIQVRR93 pKa = 11.84CGPRR97 pKa = 11.84NTFSPSHH104 pKa = 6.08FVRR107 pKa = 11.84KK108 pKa = 9.24RR109 pKa = 11.84RR110 pKa = 11.84HH111 pKa = 5.18GFLSRR116 pKa = 11.84VRR118 pKa = 11.84TRR120 pKa = 11.84KK121 pKa = 9.59GRR123 pKa = 11.84ATLQRR128 pKa = 11.84RR129 pKa = 11.84KK130 pKa = 10.21SKK132 pKa = 10.49NRR134 pKa = 11.84STLSHH139 pKa = 6.67
Molecular weight: 15.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5350055 |
51 |
5659 |
486.4 |
53.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.936 ± 0.022 | 1.161 ± 0.01 |
5.608 ± 0.014 | 6.355 ± 0.023 |
3.64 ± 0.013 | 7.306 ± 0.023 |
2.213 ± 0.01 | 5.02 ± 0.017 |
5.013 ± 0.021 | 8.769 ± 0.027 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.18 ± 0.008 | 3.709 ± 0.014 |
5.95 ± 0.022 | 3.825 ± 0.018 |
5.819 ± 0.022 | 8.072 ± 0.023 |
6.075 ± 0.016 | 6.187 ± 0.02 |
1.412 ± 0.008 | 2.748 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
