
Tenacibaculum mesophilum
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Tenacibaculum
Average proteome isoelectric point is 6.82
Get precalculated fractions of proteins
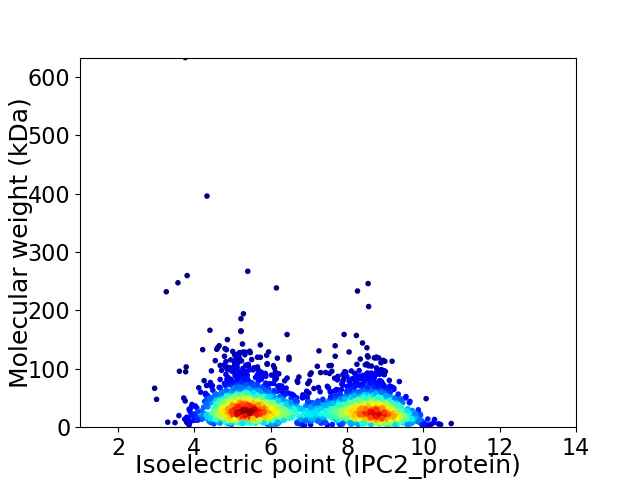
Virtual 2D-PAGE plot for 2966 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M5GT67|A0A1M5GT67_9FLAO Ribosomal large subunit pseudouridine synthase D OS=Tenacibaculum mesophilum OX=104268 GN=SAMN05444344_2568 PE=4 SV=1
MM1 pKa = 7.33KK2 pKa = 10.06NKK4 pKa = 9.64HH5 pKa = 5.23PHH7 pKa = 5.01YY8 pKa = 10.37SVYY11 pKa = 10.83KK12 pKa = 9.19SILYY16 pKa = 10.07IIFLTFTISLSSQVIIKK33 pKa = 10.47EE34 pKa = 3.85EE35 pKa = 4.04LKK37 pKa = 10.7RR38 pKa = 11.84SNLNKK43 pKa = 9.15TLSYY47 pKa = 10.59KK48 pKa = 10.45PPQDD52 pKa = 3.44TKK54 pKa = 11.23LSSSNNLVNPISSSFLTKK72 pKa = 10.53KK73 pKa = 8.59ITNSNLNQRR82 pKa = 11.84SFSSTFSMMMMATPTLDD99 pKa = 3.95LNSITPGNNYY109 pKa = 9.99DD110 pKa = 3.84FQLQPTTSNIFPVTVDD126 pKa = 3.52PKK128 pKa = 9.35VTTDD132 pKa = 3.28TGVLSATISFSGIVDD147 pKa = 3.76TPFGEE152 pKa = 4.69LLAINNGGGFDD163 pKa = 4.82LLWLQLPVSGPFTYY177 pKa = 10.07TIGSSTIQITQLTNTTFNIVEE198 pKa = 4.26TSGNPISNSEE208 pKa = 4.13FEE210 pKa = 4.7TFLSTLFYY218 pKa = 11.37GDD220 pKa = 3.94LQSPYY225 pKa = 8.63TDD227 pKa = 3.25GVRR230 pKa = 11.84TMDD233 pKa = 3.18VTIFDD238 pKa = 4.35TNGDD242 pKa = 3.77STSAQTIIRR251 pKa = 11.84IYY253 pKa = 6.69TTPPTVVDD261 pKa = 3.74EE262 pKa = 4.86ADD264 pKa = 3.91SILANSTGTITGNVLTNDD282 pKa = 3.66SGSTAISVTEE292 pKa = 3.72VDD294 pKa = 4.23VYY296 pKa = 10.95PSAVNNTYY304 pKa = 7.33TTLYY308 pKa = 11.05GDD310 pKa = 3.46ITIQANGTYY319 pKa = 9.49TYY321 pKa = 11.28DD322 pKa = 3.38VDD324 pKa = 3.71EE325 pKa = 4.87TNASVTGLRR334 pKa = 11.84NGEE337 pKa = 4.18SLDD340 pKa = 4.84DD341 pKa = 4.48IISYY345 pKa = 8.56TVADD349 pKa = 4.74AIGIKK354 pKa = 10.21DD355 pKa = 3.82YY356 pKa = 11.58GILTITINGVDD367 pKa = 4.23EE368 pKa = 5.11APDD371 pKa = 4.97AIDD374 pKa = 3.59NTDD377 pKa = 3.39SLTAFIDD384 pKa = 3.97ASATGNVITDD394 pKa = 4.2PGTGGSDD401 pKa = 3.1TVDD404 pKa = 3.12RR405 pKa = 11.84GLSTLVWEE413 pKa = 4.93SNFTNGEE420 pKa = 4.34TVNGKK425 pKa = 7.69TKK427 pKa = 10.21TIGGVGLSFTTLDD440 pKa = 3.39PGGFGTSFNQTVDD453 pKa = 3.26YY454 pKa = 7.14TTNGGHH460 pKa = 6.07TGYY463 pKa = 10.79LIYY466 pKa = 11.05NIDD469 pKa = 3.73GTTNPTDD476 pKa = 3.52DD477 pKa = 3.81TVLVIDD483 pKa = 4.49FDD485 pKa = 4.1QPVFNLGFLLTDD497 pKa = 3.35IDD499 pKa = 4.78FSQGSSWQDD508 pKa = 3.14QIKK511 pKa = 10.47IEE513 pKa = 4.24GNLDD517 pKa = 4.21GINSTYY523 pKa = 11.06KK524 pKa = 10.13FVTTGGVINSGSNTFYY540 pKa = 11.14GIGSAIPSDD549 pKa = 3.26ATGNINVFFEE559 pKa = 4.1QPINQLRR566 pKa = 11.84LSYY569 pKa = 10.96NYY571 pKa = 9.82GPNVTDD577 pKa = 5.05PDD579 pKa = 4.24PLGQIAGVSDD589 pKa = 4.53IYY591 pKa = 10.36WQGSGSVTTIRR602 pKa = 11.84LGTTLGNLNAGNLNILYY619 pKa = 8.94PGTYY623 pKa = 10.17GSIIMNPDD631 pKa = 2.44GSYY634 pKa = 10.7EE635 pKa = 4.26YY636 pKa = 10.35IVDD639 pKa = 3.91TSNPAVAGLLVGQTLTDD656 pKa = 3.35TFFYY660 pKa = 10.28EE661 pKa = 3.86ISDD664 pKa = 3.86GVSSDD669 pKa = 3.22TANLVITINGSGTDD683 pKa = 3.64PDD685 pKa = 4.21GDD687 pKa = 4.36GIADD691 pKa = 4.81RR692 pKa = 11.84LDD694 pKa = 4.55LDD696 pKa = 4.95DD697 pKa = 6.5DD698 pKa = 4.28NDD700 pKa = 5.04GILDD704 pKa = 3.96TEE706 pKa = 4.45EE707 pKa = 4.54SLACAGTPLDD717 pKa = 3.83LTGIGTTLTSGSYY730 pKa = 10.45FDD732 pKa = 4.74QDD734 pKa = 3.39TEE736 pKa = 4.55KK737 pKa = 10.68IIDD740 pKa = 3.66VTITTTGTVDD750 pKa = 2.93EE751 pKa = 5.33GNANGDD757 pKa = 3.49ILITQGGTATFTFSSPVTVSLKK779 pKa = 10.7HH780 pKa = 6.14EE781 pKa = 4.2IGITGNFDD789 pKa = 3.71TGDD792 pKa = 3.46TWEE795 pKa = 4.3LTSTGEE801 pKa = 4.13FTVADD806 pKa = 4.12PNGDD810 pKa = 3.6LMINSNSGGILNFDD824 pKa = 3.6ALGAISASEE833 pKa = 3.57AWEE836 pKa = 3.96IKK838 pKa = 7.81TTTTSLKK845 pKa = 10.65LDD847 pKa = 4.01LKK849 pKa = 10.96SGNTKK854 pKa = 10.53SPINLALICGGFLDD868 pKa = 4.82SDD870 pKa = 3.64NDD872 pKa = 4.73GIPNHH877 pKa = 7.09LDD879 pKa = 3.34LDD881 pKa = 4.22SDD883 pKa = 4.06NDD885 pKa = 3.96GCFDD889 pKa = 3.85AVEE892 pKa = 4.11ASEE895 pKa = 4.36NVSSGQLDD903 pKa = 3.71GG904 pKa = 4.71
MM1 pKa = 7.33KK2 pKa = 10.06NKK4 pKa = 9.64HH5 pKa = 5.23PHH7 pKa = 5.01YY8 pKa = 10.37SVYY11 pKa = 10.83KK12 pKa = 9.19SILYY16 pKa = 10.07IIFLTFTISLSSQVIIKK33 pKa = 10.47EE34 pKa = 3.85EE35 pKa = 4.04LKK37 pKa = 10.7RR38 pKa = 11.84SNLNKK43 pKa = 9.15TLSYY47 pKa = 10.59KK48 pKa = 10.45PPQDD52 pKa = 3.44TKK54 pKa = 11.23LSSSNNLVNPISSSFLTKK72 pKa = 10.53KK73 pKa = 8.59ITNSNLNQRR82 pKa = 11.84SFSSTFSMMMMATPTLDD99 pKa = 3.95LNSITPGNNYY109 pKa = 9.99DD110 pKa = 3.84FQLQPTTSNIFPVTVDD126 pKa = 3.52PKK128 pKa = 9.35VTTDD132 pKa = 3.28TGVLSATISFSGIVDD147 pKa = 3.76TPFGEE152 pKa = 4.69LLAINNGGGFDD163 pKa = 4.82LLWLQLPVSGPFTYY177 pKa = 10.07TIGSSTIQITQLTNTTFNIVEE198 pKa = 4.26TSGNPISNSEE208 pKa = 4.13FEE210 pKa = 4.7TFLSTLFYY218 pKa = 11.37GDD220 pKa = 3.94LQSPYY225 pKa = 8.63TDD227 pKa = 3.25GVRR230 pKa = 11.84TMDD233 pKa = 3.18VTIFDD238 pKa = 4.35TNGDD242 pKa = 3.77STSAQTIIRR251 pKa = 11.84IYY253 pKa = 6.69TTPPTVVDD261 pKa = 3.74EE262 pKa = 4.86ADD264 pKa = 3.91SILANSTGTITGNVLTNDD282 pKa = 3.66SGSTAISVTEE292 pKa = 3.72VDD294 pKa = 4.23VYY296 pKa = 10.95PSAVNNTYY304 pKa = 7.33TTLYY308 pKa = 11.05GDD310 pKa = 3.46ITIQANGTYY319 pKa = 9.49TYY321 pKa = 11.28DD322 pKa = 3.38VDD324 pKa = 3.71EE325 pKa = 4.87TNASVTGLRR334 pKa = 11.84NGEE337 pKa = 4.18SLDD340 pKa = 4.84DD341 pKa = 4.48IISYY345 pKa = 8.56TVADD349 pKa = 4.74AIGIKK354 pKa = 10.21DD355 pKa = 3.82YY356 pKa = 11.58GILTITINGVDD367 pKa = 4.23EE368 pKa = 5.11APDD371 pKa = 4.97AIDD374 pKa = 3.59NTDD377 pKa = 3.39SLTAFIDD384 pKa = 3.97ASATGNVITDD394 pKa = 4.2PGTGGSDD401 pKa = 3.1TVDD404 pKa = 3.12RR405 pKa = 11.84GLSTLVWEE413 pKa = 4.93SNFTNGEE420 pKa = 4.34TVNGKK425 pKa = 7.69TKK427 pKa = 10.21TIGGVGLSFTTLDD440 pKa = 3.39PGGFGTSFNQTVDD453 pKa = 3.26YY454 pKa = 7.14TTNGGHH460 pKa = 6.07TGYY463 pKa = 10.79LIYY466 pKa = 11.05NIDD469 pKa = 3.73GTTNPTDD476 pKa = 3.52DD477 pKa = 3.81TVLVIDD483 pKa = 4.49FDD485 pKa = 4.1QPVFNLGFLLTDD497 pKa = 3.35IDD499 pKa = 4.78FSQGSSWQDD508 pKa = 3.14QIKK511 pKa = 10.47IEE513 pKa = 4.24GNLDD517 pKa = 4.21GINSTYY523 pKa = 11.06KK524 pKa = 10.13FVTTGGVINSGSNTFYY540 pKa = 11.14GIGSAIPSDD549 pKa = 3.26ATGNINVFFEE559 pKa = 4.1QPINQLRR566 pKa = 11.84LSYY569 pKa = 10.96NYY571 pKa = 9.82GPNVTDD577 pKa = 5.05PDD579 pKa = 4.24PLGQIAGVSDD589 pKa = 4.53IYY591 pKa = 10.36WQGSGSVTTIRR602 pKa = 11.84LGTTLGNLNAGNLNILYY619 pKa = 8.94PGTYY623 pKa = 10.17GSIIMNPDD631 pKa = 2.44GSYY634 pKa = 10.7EE635 pKa = 4.26YY636 pKa = 10.35IVDD639 pKa = 3.91TSNPAVAGLLVGQTLTDD656 pKa = 3.35TFFYY660 pKa = 10.28EE661 pKa = 3.86ISDD664 pKa = 3.86GVSSDD669 pKa = 3.22TANLVITINGSGTDD683 pKa = 3.64PDD685 pKa = 4.21GDD687 pKa = 4.36GIADD691 pKa = 4.81RR692 pKa = 11.84LDD694 pKa = 4.55LDD696 pKa = 4.95DD697 pKa = 6.5DD698 pKa = 4.28NDD700 pKa = 5.04GILDD704 pKa = 3.96TEE706 pKa = 4.45EE707 pKa = 4.54SLACAGTPLDD717 pKa = 3.83LTGIGTTLTSGSYY730 pKa = 10.45FDD732 pKa = 4.74QDD734 pKa = 3.39TEE736 pKa = 4.55KK737 pKa = 10.68IIDD740 pKa = 3.66VTITTTGTVDD750 pKa = 2.93EE751 pKa = 5.33GNANGDD757 pKa = 3.49ILITQGGTATFTFSSPVTVSLKK779 pKa = 10.7HH780 pKa = 6.14EE781 pKa = 4.2IGITGNFDD789 pKa = 3.71TGDD792 pKa = 3.46TWEE795 pKa = 4.3LTSTGEE801 pKa = 4.13FTVADD806 pKa = 4.12PNGDD810 pKa = 3.6LMINSNSGGILNFDD824 pKa = 3.6ALGAISASEE833 pKa = 3.57AWEE836 pKa = 3.96IKK838 pKa = 7.81TTTTSLKK845 pKa = 10.65LDD847 pKa = 4.01LKK849 pKa = 10.96SGNTKK854 pKa = 10.53SPINLALICGGFLDD868 pKa = 4.82SDD870 pKa = 3.64NDD872 pKa = 4.73GIPNHH877 pKa = 7.09LDD879 pKa = 3.34LDD881 pKa = 4.22SDD883 pKa = 4.06NDD885 pKa = 3.96GCFDD889 pKa = 3.85AVEE892 pKa = 4.11ASEE895 pKa = 4.36NVSSGQLDD903 pKa = 3.71GG904 pKa = 4.71
Molecular weight: 95.76 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M5H9J5|A0A1M5H9J5_9FLAO Uncharacterized protein OS=Tenacibaculum mesophilum OX=104268 GN=SAMN05444344_2819 PE=4 SV=1
MM1 pKa = 7.44KK2 pKa = 10.52FIDD5 pKa = 3.93FVRR8 pKa = 11.84LFFEE12 pKa = 3.75RR13 pKa = 11.84HH14 pKa = 4.78GFNVSSRR21 pKa = 11.84FADD24 pKa = 3.65RR25 pKa = 11.84LGMRR29 pKa = 11.84ATNVRR34 pKa = 11.84LFFIYY39 pKa = 10.25ISFVTVGLSFGLYY52 pKa = 8.77LTMAFLLRR60 pKa = 11.84LKK62 pKa = 11.1DD63 pKa = 3.36MIYY66 pKa = 10.24TKK68 pKa = 9.83RR69 pKa = 11.84TSVFDD74 pKa = 3.66LL75 pKa = 4.18
MM1 pKa = 7.44KK2 pKa = 10.52FIDD5 pKa = 3.93FVRR8 pKa = 11.84LFFEE12 pKa = 3.75RR13 pKa = 11.84HH14 pKa = 4.78GFNVSSRR21 pKa = 11.84FADD24 pKa = 3.65RR25 pKa = 11.84LGMRR29 pKa = 11.84ATNVRR34 pKa = 11.84LFFIYY39 pKa = 10.25ISFVTVGLSFGLYY52 pKa = 8.77LTMAFLLRR60 pKa = 11.84LKK62 pKa = 11.1DD63 pKa = 3.36MIYY66 pKa = 10.24TKK68 pKa = 9.83RR69 pKa = 11.84TSVFDD74 pKa = 3.66LL75 pKa = 4.18
Molecular weight: 8.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
989990 |
31 |
6017 |
333.8 |
37.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.0 ± 0.049 | 0.719 ± 0.016 |
5.294 ± 0.036 | 6.859 ± 0.05 |
5.2 ± 0.039 | 6.173 ± 0.051 |
1.694 ± 0.023 | 7.995 ± 0.045 |
8.621 ± 0.067 | 9.132 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.008 ± 0.022 | 6.557 ± 0.051 |
3.205 ± 0.028 | 3.359 ± 0.027 |
3.214 ± 0.029 | 6.431 ± 0.038 |
6.089 ± 0.064 | 6.329 ± 0.038 |
0.989 ± 0.017 | 4.13 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
