
Pseudorhodobacter antarcticus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Pseudorhodobacter
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
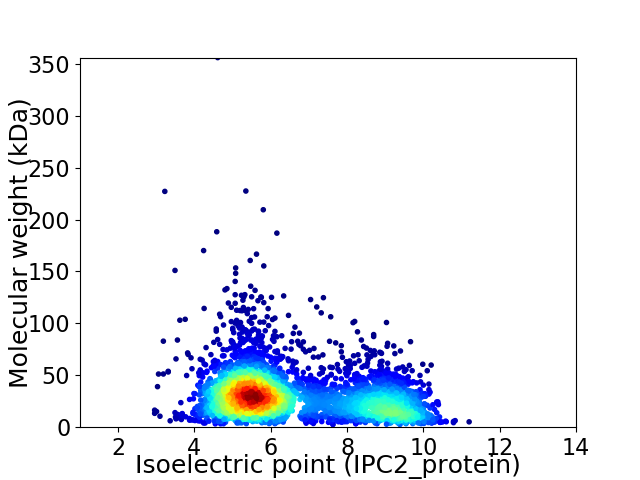
Virtual 2D-PAGE plot for 3792 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H8C4W3|A0A1H8C4W3_9RHOB Myo-inositol-1(Or 4)-monophosphatase OS=Pseudorhodobacter antarcticus OX=1077947 GN=SAMN05216227_100459 PE=3 SV=1
MM1 pKa = 7.23ATNSFDD7 pKa = 4.28NPGAQTNQTINGTATADD24 pKa = 3.61TLTGGLGNDD33 pKa = 4.39TIYY36 pKa = 11.26GGAGNDD42 pKa = 3.78FLRR45 pKa = 11.84GDD47 pKa = 3.77TGVPGSWHH55 pKa = 6.22YY56 pKa = 9.49EE57 pKa = 3.76TFNFDD62 pKa = 3.14FTSANGQAFNPAAFNTTTRR81 pKa = 11.84TGSGYY86 pKa = 8.82VTDD89 pKa = 4.5FNEE92 pKa = 4.46GGITNTMRR100 pKa = 11.84GVPASTNAEE109 pKa = 4.12DD110 pKa = 3.95FGVIYY115 pKa = 10.23TSTLNTTLGGTFRR128 pKa = 11.84LTTSSDD134 pKa = 3.42DD135 pKa = 3.49GSTVQIFNSAGVALNFNNQTGGVRR159 pKa = 11.84NYY161 pKa = 10.68LNNDD165 pKa = 3.42FHH167 pKa = 8.42QGTTTRR173 pKa = 11.84WGDD176 pKa = 3.34VVLNPNEE183 pKa = 4.2TYY185 pKa = 10.37TIQIRR190 pKa = 11.84YY191 pKa = 7.17WEE193 pKa = 4.18NRR195 pKa = 11.84GADD198 pKa = 3.73TLSATISGPATGGVQNLLTSPMLGLPPPPEE228 pKa = 3.91YY229 pKa = 10.76SVTGTPMGVQGNDD242 pKa = 3.07QLFGGDD248 pKa = 4.67GNDD251 pKa = 3.3TLYY254 pKa = 11.47GDD256 pKa = 4.89GGNDD260 pKa = 3.29TLFGGNNNDD269 pKa = 3.83LLYY272 pKa = 10.94GGTGNDD278 pKa = 3.92VLNGDD283 pKa = 4.24AGADD287 pKa = 3.47TLYY290 pKa = 11.31GDD292 pKa = 5.58DD293 pKa = 6.15GNDD296 pKa = 3.45TLNGGNDD303 pKa = 3.3NDD305 pKa = 4.68LLFGGAGADD314 pKa = 3.73SLNGDD319 pKa = 3.82VGNDD323 pKa = 3.47TLYY326 pKa = 11.54GDD328 pKa = 5.02AGNDD332 pKa = 3.6TLNGGDD338 pKa = 4.29GNDD341 pKa = 4.05LLFGGADD348 pKa = 3.37NDD350 pKa = 4.09LLYY353 pKa = 11.25GGINNDD359 pKa = 3.46TLYY362 pKa = 11.48GDD364 pKa = 4.61AGADD368 pKa = 3.41ALFGGAGNDD377 pKa = 3.67LLFGGTGNDD386 pKa = 3.73TLNGGAGNNTMTGGAGDD403 pKa = 3.87DD404 pKa = 3.77TFIYY408 pKa = 10.2TPGSTLTITDD418 pKa = 4.06FNFGNSGSITDD429 pKa = 4.51SNPSNNDD436 pKa = 2.98FLDD439 pKa = 3.85LSAYY443 pKa = 8.96YY444 pKa = 10.7ASLRR448 pKa = 11.84DD449 pKa = 3.63LRR451 pKa = 11.84NDD453 pKa = 3.4FVDD456 pKa = 5.77DD457 pKa = 5.1GILNQSVGNYY467 pKa = 6.06TTTTALGGTITLTGISATDD486 pKa = 3.45LTRR489 pKa = 11.84DD490 pKa = 4.07NINVACFTLGTLIDD504 pKa = 3.93TATGPRR510 pKa = 11.84AIEE513 pKa = 3.82TLAPGTLVCTADD525 pKa = 4.45HH526 pKa = 6.67GLQPLRR532 pKa = 11.84AVLSRR537 pKa = 11.84DD538 pKa = 3.25VPGDD542 pKa = 3.44GRR544 pKa = 11.84HH545 pKa = 5.42APVMFRR551 pKa = 11.84AGALGNTRR559 pKa = 11.84DD560 pKa = 3.88MLTSPAHH567 pKa = 6.56RR568 pKa = 11.84MMITDD573 pKa = 3.7WRR575 pKa = 11.84AEE577 pKa = 3.84LLFGEE582 pKa = 5.02GEE584 pKa = 4.42VLVTARR590 pKa = 11.84ALLNDD595 pKa = 3.82HH596 pKa = 6.99SITRR600 pKa = 11.84APCARR605 pKa = 11.84VTYY608 pKa = 9.88YY609 pKa = 10.88HH610 pKa = 7.16LLLDD614 pKa = 3.59SHH616 pKa = 6.87EE617 pKa = 4.13IVYY620 pKa = 10.99AEE622 pKa = 5.01GIATEE627 pKa = 4.81SYY629 pKa = 9.09HH630 pKa = 7.06HH631 pKa = 6.48SAAVADD637 pKa = 4.11AEE639 pKa = 4.75VAAEE643 pKa = 3.93LHH645 pKa = 6.87ALFPDD650 pKa = 4.48LAQSKK655 pKa = 7.99TPSARR660 pKa = 11.84LSLRR664 pKa = 11.84GYY666 pKa = 9.51EE667 pKa = 4.07AATLLAAAA675 pKa = 5.25
MM1 pKa = 7.23ATNSFDD7 pKa = 4.28NPGAQTNQTINGTATADD24 pKa = 3.61TLTGGLGNDD33 pKa = 4.39TIYY36 pKa = 11.26GGAGNDD42 pKa = 3.78FLRR45 pKa = 11.84GDD47 pKa = 3.77TGVPGSWHH55 pKa = 6.22YY56 pKa = 9.49EE57 pKa = 3.76TFNFDD62 pKa = 3.14FTSANGQAFNPAAFNTTTRR81 pKa = 11.84TGSGYY86 pKa = 8.82VTDD89 pKa = 4.5FNEE92 pKa = 4.46GGITNTMRR100 pKa = 11.84GVPASTNAEE109 pKa = 4.12DD110 pKa = 3.95FGVIYY115 pKa = 10.23TSTLNTTLGGTFRR128 pKa = 11.84LTTSSDD134 pKa = 3.42DD135 pKa = 3.49GSTVQIFNSAGVALNFNNQTGGVRR159 pKa = 11.84NYY161 pKa = 10.68LNNDD165 pKa = 3.42FHH167 pKa = 8.42QGTTTRR173 pKa = 11.84WGDD176 pKa = 3.34VVLNPNEE183 pKa = 4.2TYY185 pKa = 10.37TIQIRR190 pKa = 11.84YY191 pKa = 7.17WEE193 pKa = 4.18NRR195 pKa = 11.84GADD198 pKa = 3.73TLSATISGPATGGVQNLLTSPMLGLPPPPEE228 pKa = 3.91YY229 pKa = 10.76SVTGTPMGVQGNDD242 pKa = 3.07QLFGGDD248 pKa = 4.67GNDD251 pKa = 3.3TLYY254 pKa = 11.47GDD256 pKa = 4.89GGNDD260 pKa = 3.29TLFGGNNNDD269 pKa = 3.83LLYY272 pKa = 10.94GGTGNDD278 pKa = 3.92VLNGDD283 pKa = 4.24AGADD287 pKa = 3.47TLYY290 pKa = 11.31GDD292 pKa = 5.58DD293 pKa = 6.15GNDD296 pKa = 3.45TLNGGNDD303 pKa = 3.3NDD305 pKa = 4.68LLFGGAGADD314 pKa = 3.73SLNGDD319 pKa = 3.82VGNDD323 pKa = 3.47TLYY326 pKa = 11.54GDD328 pKa = 5.02AGNDD332 pKa = 3.6TLNGGDD338 pKa = 4.29GNDD341 pKa = 4.05LLFGGADD348 pKa = 3.37NDD350 pKa = 4.09LLYY353 pKa = 11.25GGINNDD359 pKa = 3.46TLYY362 pKa = 11.48GDD364 pKa = 4.61AGADD368 pKa = 3.41ALFGGAGNDD377 pKa = 3.67LLFGGTGNDD386 pKa = 3.73TLNGGAGNNTMTGGAGDD403 pKa = 3.87DD404 pKa = 3.77TFIYY408 pKa = 10.2TPGSTLTITDD418 pKa = 4.06FNFGNSGSITDD429 pKa = 4.51SNPSNNDD436 pKa = 2.98FLDD439 pKa = 3.85LSAYY443 pKa = 8.96YY444 pKa = 10.7ASLRR448 pKa = 11.84DD449 pKa = 3.63LRR451 pKa = 11.84NDD453 pKa = 3.4FVDD456 pKa = 5.77DD457 pKa = 5.1GILNQSVGNYY467 pKa = 6.06TTTTALGGTITLTGISATDD486 pKa = 3.45LTRR489 pKa = 11.84DD490 pKa = 4.07NINVACFTLGTLIDD504 pKa = 3.93TATGPRR510 pKa = 11.84AIEE513 pKa = 3.82TLAPGTLVCTADD525 pKa = 4.45HH526 pKa = 6.67GLQPLRR532 pKa = 11.84AVLSRR537 pKa = 11.84DD538 pKa = 3.25VPGDD542 pKa = 3.44GRR544 pKa = 11.84HH545 pKa = 5.42APVMFRR551 pKa = 11.84AGALGNTRR559 pKa = 11.84DD560 pKa = 3.88MLTSPAHH567 pKa = 6.56RR568 pKa = 11.84MMITDD573 pKa = 3.7WRR575 pKa = 11.84AEE577 pKa = 3.84LLFGEE582 pKa = 5.02GEE584 pKa = 4.42VLVTARR590 pKa = 11.84ALLNDD595 pKa = 3.82HH596 pKa = 6.99SITRR600 pKa = 11.84APCARR605 pKa = 11.84VTYY608 pKa = 9.88YY609 pKa = 10.88HH610 pKa = 7.16LLLDD614 pKa = 3.59SHH616 pKa = 6.87EE617 pKa = 4.13IVYY620 pKa = 10.99AEE622 pKa = 5.01GIATEE627 pKa = 4.81SYY629 pKa = 9.09HH630 pKa = 7.06HH631 pKa = 6.48SAAVADD637 pKa = 4.11AEE639 pKa = 4.75VAAEE643 pKa = 3.93LHH645 pKa = 6.87ALFPDD650 pKa = 4.48LAQSKK655 pKa = 7.99TPSARR660 pKa = 11.84LSLRR664 pKa = 11.84GYY666 pKa = 9.51EE667 pKa = 4.07AATLLAAAA675 pKa = 5.25
Molecular weight: 70.07 kDa
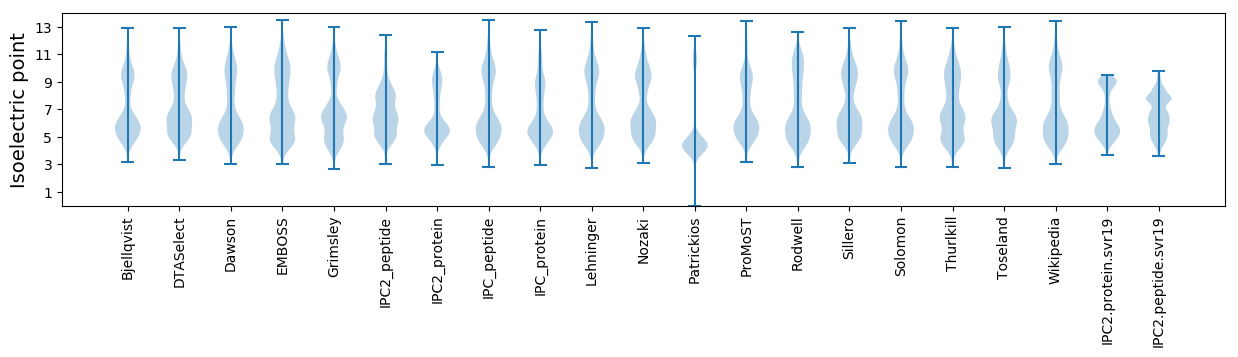
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H8FJF0|A0A1H8FJF0_9RHOB Bifunctional purine biosynthesis protein PurH OS=Pseudorhodobacter antarcticus OX=1077947 GN=purH PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84LRR21 pKa = 11.84MATKK25 pKa = 10.34AGRR28 pKa = 11.84NILNLRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.16GRR39 pKa = 11.84KK40 pKa = 8.85KK41 pKa = 10.63LSAA44 pKa = 3.95
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84LRR21 pKa = 11.84MATKK25 pKa = 10.34AGRR28 pKa = 11.84NILNLRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.16GRR39 pKa = 11.84KK40 pKa = 8.85KK41 pKa = 10.63LSAA44 pKa = 3.95
Molecular weight: 5.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1132519 |
24 |
3262 |
298.7 |
32.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.091 ± 0.068 | 0.903 ± 0.013 |
5.912 ± 0.036 | 4.766 ± 0.037 |
3.772 ± 0.023 | 8.831 ± 0.054 |
2.078 ± 0.025 | 5.244 ± 0.026 |
3.185 ± 0.035 | 10.067 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.972 ± 0.019 | 2.7 ± 0.026 |
5.112 ± 0.034 | 3.292 ± 0.022 |
6.44 ± 0.037 | 4.851 ± 0.027 |
5.816 ± 0.029 | 7.402 ± 0.041 |
1.408 ± 0.018 | 2.158 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
