
Labrenzia suaedae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Stappiaceae; Labrenzia
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
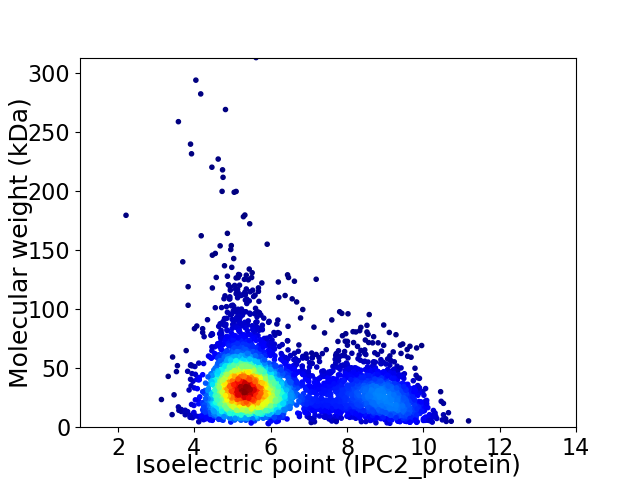
Virtual 2D-PAGE plot for 4539 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
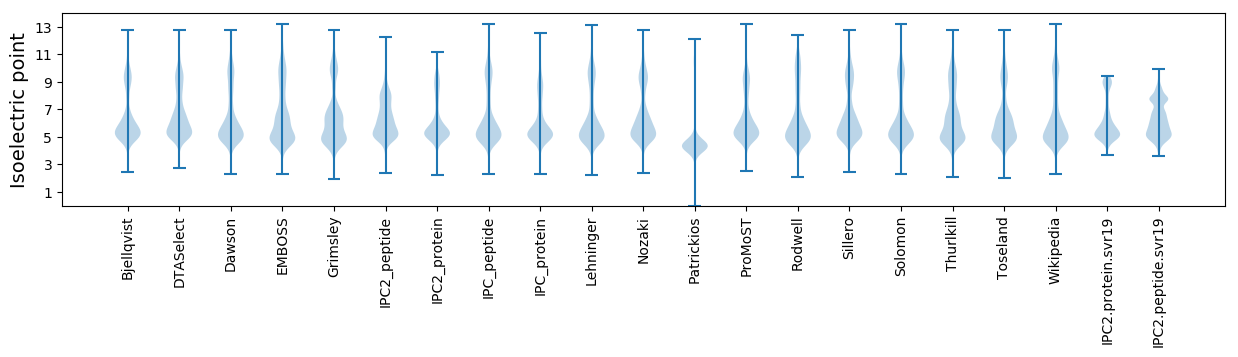
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M6YXA1|A0A1M6YXA1_9RHOB Putative peptidoglycan binding domain-containing protein OS=Labrenzia suaedae OX=735517 GN=SAMN05444272_0062 PE=4 SV=1
MM1 pKa = 7.24VKK3 pKa = 10.67NSIDD7 pKa = 3.04WGTQVAASGGVINVYY22 pKa = 9.57FAEE25 pKa = 4.34SGEE28 pKa = 4.26VFDD31 pKa = 5.43GQTSQGWSSYY41 pKa = 10.25EE42 pKa = 3.53IQQAMLAFSSYY53 pKa = 11.7SNFLNITFNRR63 pKa = 11.84VYY65 pKa = 10.42SSSEE69 pKa = 3.51ATFNLVATSSSSIGGNLGYY88 pKa = 9.59MNPPGTSGAGIGVFATDD105 pKa = 3.35GTGWNTYY112 pKa = 10.0GGLEE116 pKa = 4.06VGGYY120 pKa = 10.66GYY122 pKa = 8.44ITLIHH127 pKa = 6.32EE128 pKa = 5.34FGHH131 pKa = 6.28GMGLAHH137 pKa = 6.82PHH139 pKa = 6.08DD140 pKa = 4.49TGGSSSIMSGVTASFGDD157 pKa = 3.83YY158 pKa = 11.28GDD160 pKa = 4.8FDD162 pKa = 5.68LNQGVYY168 pKa = 8.92TAMSYY173 pKa = 11.16NDD175 pKa = 3.29GWQTAPHH182 pKa = 6.44GASPSYY188 pKa = 10.69SYY190 pKa = 10.99GYY192 pKa = 9.27EE193 pKa = 3.99GTMMALDD200 pKa = 3.46IAVLQDD206 pKa = 3.2KK207 pKa = 10.72YY208 pKa = 11.42GANTTFASGNNTYY221 pKa = 10.66VLPGSNGVGTYY232 pKa = 10.22YY233 pKa = 10.95SSIWDD238 pKa = 3.32TGGNDD243 pKa = 3.51EE244 pKa = 4.74ISYY247 pKa = 9.48TGSLDD252 pKa = 3.25VTIDD256 pKa = 3.84LRR258 pKa = 11.84PATLNYY264 pKa = 9.88EE265 pKa = 4.13VGGGGFISYY274 pKa = 10.88ASGIHH279 pKa = 5.91GGYY282 pKa = 8.98TIAQGVVIEE291 pKa = 4.69FATSGSGNDD300 pKa = 3.6TLVGNAANNVLRR312 pKa = 11.84GNGGNDD318 pKa = 2.98TLTGNRR324 pKa = 11.84GNDD327 pKa = 3.44KK328 pKa = 10.47IYY330 pKa = 10.82GGTGSDD336 pKa = 3.4VAVYY340 pKa = 10.33NYY342 pKa = 10.45NSSEE346 pKa = 3.89YY347 pKa = 10.25TITHH351 pKa = 6.64NDD353 pKa = 3.64DD354 pKa = 3.26GSITIAHH361 pKa = 6.3TGGTRR366 pKa = 11.84LDD368 pKa = 4.25GVDD371 pKa = 3.11TLYY374 pKa = 11.04DD375 pKa = 3.36VEE377 pKa = 4.36AAQFADD383 pKa = 3.56EE384 pKa = 5.69RR385 pKa = 11.84IVLDD389 pKa = 4.29KK390 pKa = 10.82LTGPLDD396 pKa = 3.69VVFLQDD402 pKa = 3.43LTGSFDD408 pKa = 4.43DD409 pKa = 5.35DD410 pKa = 4.06LPYY413 pKa = 9.87MQNSVDD419 pKa = 5.29DD420 pKa = 3.74IAEE423 pKa = 4.34SIEE426 pKa = 3.98LSFSGSRR433 pKa = 11.84FAVTSFKK440 pKa = 10.74DD441 pKa = 2.93IYY443 pKa = 11.12DD444 pKa = 3.81EE445 pKa = 3.77YY446 pKa = 10.56TYY448 pKa = 11.29AVEE451 pKa = 5.27ANFTTSTASLSSTYY465 pKa = 10.93AGLSASGGGDD475 pKa = 3.09SAEE478 pKa = 4.24AQLTALLSAANDD490 pKa = 3.55TTLSYY495 pKa = 10.59RR496 pKa = 11.84VGVSRR501 pKa = 11.84VFVLATDD508 pKa = 4.41ASFHH512 pKa = 6.6SYY514 pKa = 10.67EE515 pKa = 4.41SIEE518 pKa = 4.28TIAATFAEE526 pKa = 4.71KK527 pKa = 10.55DD528 pKa = 3.53IIPIFAVTSSQVGTYY543 pKa = 9.78QGLVDD548 pKa = 4.25QIGAGIVVTIQYY560 pKa = 10.18DD561 pKa = 3.44SSDD564 pKa = 3.57FADD567 pKa = 4.01SIRR570 pKa = 11.84YY571 pKa = 8.42GLAEE575 pKa = 4.19LSGEE579 pKa = 4.22VTEE582 pKa = 6.03LGTSSNDD589 pKa = 2.65ILTGKK594 pKa = 10.09EE595 pKa = 4.0GFSDD599 pKa = 4.61YY600 pKa = 10.94IFGMAGDD607 pKa = 3.87DD608 pKa = 4.13SIRR611 pKa = 11.84GLGGNDD617 pKa = 3.4KK618 pKa = 11.04LDD620 pKa = 4.06GGSGDD625 pKa = 5.09DD626 pKa = 5.02DD627 pKa = 5.22LSGDD631 pKa = 3.95SGDD634 pKa = 3.74DD635 pKa = 3.54TVIGGSGKK643 pKa = 10.18DD644 pKa = 3.38VLRR647 pKa = 11.84GGTGSDD653 pKa = 4.69KK654 pKa = 10.75IDD656 pKa = 3.68GNQGDD661 pKa = 4.1DD662 pKa = 3.38TLYY665 pKa = 11.21GDD667 pKa = 5.45DD668 pKa = 4.07VGTTSGFGNDD678 pKa = 4.0EE679 pKa = 4.41LNGGDD684 pKa = 4.21GNDD687 pKa = 3.57ILVGDD692 pKa = 4.97LGSDD696 pKa = 3.45KK697 pKa = 11.08LDD699 pKa = 3.83GGAGSDD705 pKa = 2.63WVYY708 pKa = 10.88YY709 pKa = 10.22QNSTSAIRR717 pKa = 11.84VDD719 pKa = 4.12LSSRR723 pKa = 11.84AAASGGYY730 pKa = 9.59AQGDD734 pKa = 3.81VISNIEE740 pKa = 4.34NILGTRR746 pKa = 11.84FSDD749 pKa = 4.31TIAGNGSANFLRR761 pKa = 11.84GGNSGDD767 pKa = 3.67TLIGNAGNDD776 pKa = 4.19TLWGEE781 pKa = 4.08QGNDD785 pKa = 3.12ILRR788 pKa = 11.84GDD790 pKa = 4.01AGADD794 pKa = 3.23ILNGGAGFDD803 pKa = 3.04WAYY806 pKa = 11.22YY807 pKa = 9.09HH808 pKa = 7.15LSDD811 pKa = 3.67AAVRR815 pKa = 11.84IDD817 pKa = 3.69LAGGAVQHH825 pKa = 6.53GGHH828 pKa = 6.87AEE830 pKa = 3.91GDD832 pKa = 3.75TLIGIEE838 pKa = 4.33NILGSVHH845 pKa = 7.63DD846 pKa = 5.97DD847 pKa = 3.42IITGNTGANHH857 pKa = 6.44LRR859 pKa = 11.84GHH861 pKa = 7.51LGNDD865 pKa = 3.21SLIGNEE871 pKa = 4.85GDD873 pKa = 3.4DD874 pKa = 4.22TLWGEE879 pKa = 4.35AGNDD883 pKa = 3.62VLRR886 pKa = 11.84GDD888 pKa = 4.22AGADD892 pKa = 3.49VLDD895 pKa = 5.02GGSGDD900 pKa = 3.44DD901 pKa = 2.9WAYY904 pKa = 9.73YY905 pKa = 9.49QKK907 pKa = 11.08SDD909 pKa = 3.27AAVVVDD915 pKa = 5.13LSDD918 pKa = 3.58GATEE922 pKa = 4.32QGGHH926 pKa = 5.6AQGDD930 pKa = 3.97TLISIEE936 pKa = 4.25RR937 pKa = 11.84VLGSTFGDD945 pKa = 4.0SITGNAGANFLRR957 pKa = 11.84GFDD960 pKa = 4.3GNDD963 pKa = 3.13NLIGNAGDD971 pKa = 3.57DD972 pKa = 4.21VLWGEE977 pKa = 4.6AGNDD981 pKa = 3.34ILRR984 pKa = 11.84GDD986 pKa = 4.26AGADD990 pKa = 3.56TLDD993 pKa = 4.19GGAGSDD999 pKa = 2.71WAYY1002 pKa = 10.31YY1003 pKa = 9.57QNSNAAINVNMADD1016 pKa = 3.77RR1017 pKa = 11.84AAEE1020 pKa = 4.27TGGHH1024 pKa = 5.54AQGDD1028 pKa = 3.94VLSNIEE1034 pKa = 4.08NVLGSFFADD1043 pKa = 4.78TIAGNAGANFLRR1055 pKa = 11.84GFNSGDD1061 pKa = 3.49MLIGNAGNDD1070 pKa = 3.82ILWGEE1075 pKa = 4.16QGNDD1079 pKa = 3.12ILRR1082 pKa = 11.84GDD1084 pKa = 3.94AGADD1088 pKa = 3.28VLNGGEE1094 pKa = 4.47GYY1096 pKa = 10.76DD1097 pKa = 2.97WAYY1100 pKa = 11.26YY1101 pKa = 9.58HH1102 pKa = 7.03LSDD1105 pKa = 4.77AAVSVDD1111 pKa = 3.68LTQGAVQHH1119 pKa = 6.42GGHH1122 pKa = 6.87AEE1124 pKa = 4.05GDD1126 pKa = 3.57TLIAIEE1132 pKa = 4.35HH1133 pKa = 5.78VLGSVHH1139 pKa = 7.52DD1140 pKa = 4.56DD1141 pKa = 3.86SIIGNSGANHH1151 pKa = 6.26LRR1153 pKa = 11.84GHH1155 pKa = 7.5LGNDD1159 pKa = 3.2SLVGNGGDD1167 pKa = 3.85DD1168 pKa = 3.8ILWGEE1173 pKa = 4.49AGDD1176 pKa = 4.11DD1177 pKa = 3.91VLRR1180 pKa = 11.84GDD1182 pKa = 4.63AGSDD1186 pKa = 3.37VLNGGTGNDD1195 pKa = 2.79WAYY1198 pKa = 10.09YY1199 pKa = 8.7QKK1201 pKa = 10.98SDD1203 pKa = 3.5AAIIVNLLDD1212 pKa = 3.87GLSEE1216 pKa = 4.38HH1217 pKa = 6.9GGHH1220 pKa = 7.31AEE1222 pKa = 3.81GDD1224 pKa = 3.71TLIGIEE1230 pKa = 4.11RR1231 pKa = 11.84VLGSLYY1237 pKa = 10.62DD1238 pKa = 4.23DD1239 pKa = 5.89SITGDD1244 pKa = 3.19NNANFLRR1251 pKa = 11.84GFNGNDD1257 pKa = 3.14TLVGNGGDD1265 pKa = 3.86DD1266 pKa = 3.71ILWGEE1271 pKa = 4.75LGTDD1275 pKa = 3.52TLTGGTGKK1283 pKa = 8.91DD1284 pKa = 3.19TFHH1287 pKa = 7.4FIADD1291 pKa = 4.07GATDD1295 pKa = 3.71TVTDD1299 pKa = 4.13FEE1301 pKa = 5.78DD1302 pKa = 5.98GIDD1305 pKa = 3.73LLSINWGATSLSQLAISEE1323 pKa = 4.37TGAGNVDD1330 pKa = 3.55TLISFGGNSILLQNFDD1346 pKa = 3.89HH1347 pKa = 6.79TLLTTDD1353 pKa = 4.2DD1354 pKa = 3.78FRR1356 pKa = 11.84FVV1358 pKa = 3.16
MM1 pKa = 7.24VKK3 pKa = 10.67NSIDD7 pKa = 3.04WGTQVAASGGVINVYY22 pKa = 9.57FAEE25 pKa = 4.34SGEE28 pKa = 4.26VFDD31 pKa = 5.43GQTSQGWSSYY41 pKa = 10.25EE42 pKa = 3.53IQQAMLAFSSYY53 pKa = 11.7SNFLNITFNRR63 pKa = 11.84VYY65 pKa = 10.42SSSEE69 pKa = 3.51ATFNLVATSSSSIGGNLGYY88 pKa = 9.59MNPPGTSGAGIGVFATDD105 pKa = 3.35GTGWNTYY112 pKa = 10.0GGLEE116 pKa = 4.06VGGYY120 pKa = 10.66GYY122 pKa = 8.44ITLIHH127 pKa = 6.32EE128 pKa = 5.34FGHH131 pKa = 6.28GMGLAHH137 pKa = 6.82PHH139 pKa = 6.08DD140 pKa = 4.49TGGSSSIMSGVTASFGDD157 pKa = 3.83YY158 pKa = 11.28GDD160 pKa = 4.8FDD162 pKa = 5.68LNQGVYY168 pKa = 8.92TAMSYY173 pKa = 11.16NDD175 pKa = 3.29GWQTAPHH182 pKa = 6.44GASPSYY188 pKa = 10.69SYY190 pKa = 10.99GYY192 pKa = 9.27EE193 pKa = 3.99GTMMALDD200 pKa = 3.46IAVLQDD206 pKa = 3.2KK207 pKa = 10.72YY208 pKa = 11.42GANTTFASGNNTYY221 pKa = 10.66VLPGSNGVGTYY232 pKa = 10.22YY233 pKa = 10.95SSIWDD238 pKa = 3.32TGGNDD243 pKa = 3.51EE244 pKa = 4.74ISYY247 pKa = 9.48TGSLDD252 pKa = 3.25VTIDD256 pKa = 3.84LRR258 pKa = 11.84PATLNYY264 pKa = 9.88EE265 pKa = 4.13VGGGGFISYY274 pKa = 10.88ASGIHH279 pKa = 5.91GGYY282 pKa = 8.98TIAQGVVIEE291 pKa = 4.69FATSGSGNDD300 pKa = 3.6TLVGNAANNVLRR312 pKa = 11.84GNGGNDD318 pKa = 2.98TLTGNRR324 pKa = 11.84GNDD327 pKa = 3.44KK328 pKa = 10.47IYY330 pKa = 10.82GGTGSDD336 pKa = 3.4VAVYY340 pKa = 10.33NYY342 pKa = 10.45NSSEE346 pKa = 3.89YY347 pKa = 10.25TITHH351 pKa = 6.64NDD353 pKa = 3.64DD354 pKa = 3.26GSITIAHH361 pKa = 6.3TGGTRR366 pKa = 11.84LDD368 pKa = 4.25GVDD371 pKa = 3.11TLYY374 pKa = 11.04DD375 pKa = 3.36VEE377 pKa = 4.36AAQFADD383 pKa = 3.56EE384 pKa = 5.69RR385 pKa = 11.84IVLDD389 pKa = 4.29KK390 pKa = 10.82LTGPLDD396 pKa = 3.69VVFLQDD402 pKa = 3.43LTGSFDD408 pKa = 4.43DD409 pKa = 5.35DD410 pKa = 4.06LPYY413 pKa = 9.87MQNSVDD419 pKa = 5.29DD420 pKa = 3.74IAEE423 pKa = 4.34SIEE426 pKa = 3.98LSFSGSRR433 pKa = 11.84FAVTSFKK440 pKa = 10.74DD441 pKa = 2.93IYY443 pKa = 11.12DD444 pKa = 3.81EE445 pKa = 3.77YY446 pKa = 10.56TYY448 pKa = 11.29AVEE451 pKa = 5.27ANFTTSTASLSSTYY465 pKa = 10.93AGLSASGGGDD475 pKa = 3.09SAEE478 pKa = 4.24AQLTALLSAANDD490 pKa = 3.55TTLSYY495 pKa = 10.59RR496 pKa = 11.84VGVSRR501 pKa = 11.84VFVLATDD508 pKa = 4.41ASFHH512 pKa = 6.6SYY514 pKa = 10.67EE515 pKa = 4.41SIEE518 pKa = 4.28TIAATFAEE526 pKa = 4.71KK527 pKa = 10.55DD528 pKa = 3.53IIPIFAVTSSQVGTYY543 pKa = 9.78QGLVDD548 pKa = 4.25QIGAGIVVTIQYY560 pKa = 10.18DD561 pKa = 3.44SSDD564 pKa = 3.57FADD567 pKa = 4.01SIRR570 pKa = 11.84YY571 pKa = 8.42GLAEE575 pKa = 4.19LSGEE579 pKa = 4.22VTEE582 pKa = 6.03LGTSSNDD589 pKa = 2.65ILTGKK594 pKa = 10.09EE595 pKa = 4.0GFSDD599 pKa = 4.61YY600 pKa = 10.94IFGMAGDD607 pKa = 3.87DD608 pKa = 4.13SIRR611 pKa = 11.84GLGGNDD617 pKa = 3.4KK618 pKa = 11.04LDD620 pKa = 4.06GGSGDD625 pKa = 5.09DD626 pKa = 5.02DD627 pKa = 5.22LSGDD631 pKa = 3.95SGDD634 pKa = 3.74DD635 pKa = 3.54TVIGGSGKK643 pKa = 10.18DD644 pKa = 3.38VLRR647 pKa = 11.84GGTGSDD653 pKa = 4.69KK654 pKa = 10.75IDD656 pKa = 3.68GNQGDD661 pKa = 4.1DD662 pKa = 3.38TLYY665 pKa = 11.21GDD667 pKa = 5.45DD668 pKa = 4.07VGTTSGFGNDD678 pKa = 4.0EE679 pKa = 4.41LNGGDD684 pKa = 4.21GNDD687 pKa = 3.57ILVGDD692 pKa = 4.97LGSDD696 pKa = 3.45KK697 pKa = 11.08LDD699 pKa = 3.83GGAGSDD705 pKa = 2.63WVYY708 pKa = 10.88YY709 pKa = 10.22QNSTSAIRR717 pKa = 11.84VDD719 pKa = 4.12LSSRR723 pKa = 11.84AAASGGYY730 pKa = 9.59AQGDD734 pKa = 3.81VISNIEE740 pKa = 4.34NILGTRR746 pKa = 11.84FSDD749 pKa = 4.31TIAGNGSANFLRR761 pKa = 11.84GGNSGDD767 pKa = 3.67TLIGNAGNDD776 pKa = 4.19TLWGEE781 pKa = 4.08QGNDD785 pKa = 3.12ILRR788 pKa = 11.84GDD790 pKa = 4.01AGADD794 pKa = 3.23ILNGGAGFDD803 pKa = 3.04WAYY806 pKa = 11.22YY807 pKa = 9.09HH808 pKa = 7.15LSDD811 pKa = 3.67AAVRR815 pKa = 11.84IDD817 pKa = 3.69LAGGAVQHH825 pKa = 6.53GGHH828 pKa = 6.87AEE830 pKa = 3.91GDD832 pKa = 3.75TLIGIEE838 pKa = 4.33NILGSVHH845 pKa = 7.63DD846 pKa = 5.97DD847 pKa = 3.42IITGNTGANHH857 pKa = 6.44LRR859 pKa = 11.84GHH861 pKa = 7.51LGNDD865 pKa = 3.21SLIGNEE871 pKa = 4.85GDD873 pKa = 3.4DD874 pKa = 4.22TLWGEE879 pKa = 4.35AGNDD883 pKa = 3.62VLRR886 pKa = 11.84GDD888 pKa = 4.22AGADD892 pKa = 3.49VLDD895 pKa = 5.02GGSGDD900 pKa = 3.44DD901 pKa = 2.9WAYY904 pKa = 9.73YY905 pKa = 9.49QKK907 pKa = 11.08SDD909 pKa = 3.27AAVVVDD915 pKa = 5.13LSDD918 pKa = 3.58GATEE922 pKa = 4.32QGGHH926 pKa = 5.6AQGDD930 pKa = 3.97TLISIEE936 pKa = 4.25RR937 pKa = 11.84VLGSTFGDD945 pKa = 4.0SITGNAGANFLRR957 pKa = 11.84GFDD960 pKa = 4.3GNDD963 pKa = 3.13NLIGNAGDD971 pKa = 3.57DD972 pKa = 4.21VLWGEE977 pKa = 4.6AGNDD981 pKa = 3.34ILRR984 pKa = 11.84GDD986 pKa = 4.26AGADD990 pKa = 3.56TLDD993 pKa = 4.19GGAGSDD999 pKa = 2.71WAYY1002 pKa = 10.31YY1003 pKa = 9.57QNSNAAINVNMADD1016 pKa = 3.77RR1017 pKa = 11.84AAEE1020 pKa = 4.27TGGHH1024 pKa = 5.54AQGDD1028 pKa = 3.94VLSNIEE1034 pKa = 4.08NVLGSFFADD1043 pKa = 4.78TIAGNAGANFLRR1055 pKa = 11.84GFNSGDD1061 pKa = 3.49MLIGNAGNDD1070 pKa = 3.82ILWGEE1075 pKa = 4.16QGNDD1079 pKa = 3.12ILRR1082 pKa = 11.84GDD1084 pKa = 3.94AGADD1088 pKa = 3.28VLNGGEE1094 pKa = 4.47GYY1096 pKa = 10.76DD1097 pKa = 2.97WAYY1100 pKa = 11.26YY1101 pKa = 9.58HH1102 pKa = 7.03LSDD1105 pKa = 4.77AAVSVDD1111 pKa = 3.68LTQGAVQHH1119 pKa = 6.42GGHH1122 pKa = 6.87AEE1124 pKa = 4.05GDD1126 pKa = 3.57TLIAIEE1132 pKa = 4.35HH1133 pKa = 5.78VLGSVHH1139 pKa = 7.52DD1140 pKa = 4.56DD1141 pKa = 3.86SIIGNSGANHH1151 pKa = 6.26LRR1153 pKa = 11.84GHH1155 pKa = 7.5LGNDD1159 pKa = 3.2SLVGNGGDD1167 pKa = 3.85DD1168 pKa = 3.8ILWGEE1173 pKa = 4.49AGDD1176 pKa = 4.11DD1177 pKa = 3.91VLRR1180 pKa = 11.84GDD1182 pKa = 4.63AGSDD1186 pKa = 3.37VLNGGTGNDD1195 pKa = 2.79WAYY1198 pKa = 10.09YY1199 pKa = 8.7QKK1201 pKa = 10.98SDD1203 pKa = 3.5AAIIVNLLDD1212 pKa = 3.87GLSEE1216 pKa = 4.38HH1217 pKa = 6.9GGHH1220 pKa = 7.31AEE1222 pKa = 3.81GDD1224 pKa = 3.71TLIGIEE1230 pKa = 4.11RR1231 pKa = 11.84VLGSLYY1237 pKa = 10.62DD1238 pKa = 4.23DD1239 pKa = 5.89SITGDD1244 pKa = 3.19NNANFLRR1251 pKa = 11.84GFNGNDD1257 pKa = 3.14TLVGNGGDD1265 pKa = 3.86DD1266 pKa = 3.71ILWGEE1271 pKa = 4.75LGTDD1275 pKa = 3.52TLTGGTGKK1283 pKa = 8.91DD1284 pKa = 3.19TFHH1287 pKa = 7.4FIADD1291 pKa = 4.07GATDD1295 pKa = 3.71TVTDD1299 pKa = 4.13FEE1301 pKa = 5.78DD1302 pKa = 5.98GIDD1305 pKa = 3.73LLSINWGATSLSQLAISEE1323 pKa = 4.37TGAGNVDD1330 pKa = 3.55TLISFGGNSILLQNFDD1346 pKa = 3.89HH1347 pKa = 6.79TLLTTDD1353 pKa = 4.2DD1354 pKa = 3.78FRR1356 pKa = 11.84FVV1358 pKa = 3.16
Molecular weight: 140.16 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M7AFC1|A0A1M7AFC1_9RHOB Putrescine-binding periplasmic protein OS=Labrenzia suaedae OX=735517 GN=SAMN05444272_0526 PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.2QPSRR9 pKa = 11.84LVRR12 pKa = 11.84KK13 pKa = 8.99RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.7GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.48NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.2QPSRR9 pKa = 11.84LVRR12 pKa = 11.84KK13 pKa = 8.99RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.7GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.48NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1505445 |
29 |
2916 |
331.7 |
35.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.586 ± 0.049 | 0.867 ± 0.011 |
5.7 ± 0.039 | 5.991 ± 0.04 |
3.886 ± 0.023 | 8.571 ± 0.054 |
1.919 ± 0.017 | 5.35 ± 0.03 |
3.761 ± 0.029 | 10.361 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.553 ± 0.019 | 2.792 ± 0.025 |
4.886 ± 0.033 | 3.275 ± 0.022 |
6.2 ± 0.045 | 6.155 ± 0.036 |
5.472 ± 0.045 | 7.169 ± 0.025 |
1.258 ± 0.015 | 2.249 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
