
Planococcus halotolerans
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Planococcaceae; Planococcus
Average proteome isoelectric point is 5.91
Get precalculated fractions of proteins
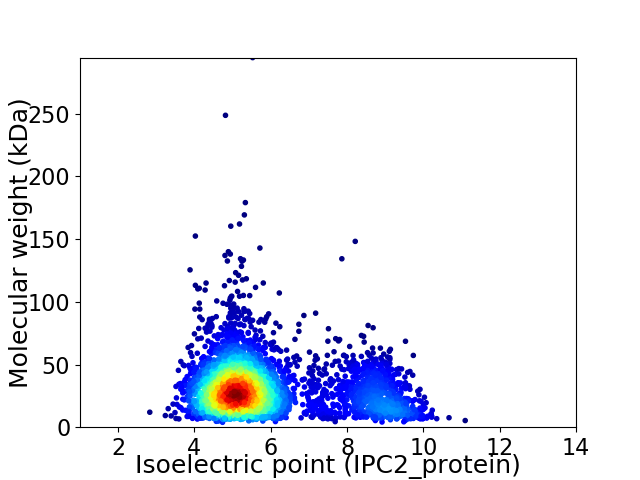
Virtual 2D-PAGE plot for 3547 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A365KRL2|A0A365KRL2_9BACL Transcriptional regulator OS=Planococcus halotolerans OX=2233542 GN=DP120_13310 PE=4 SV=1
MM1 pKa = 7.42KK2 pKa = 10.13KK3 pKa = 10.15KK4 pKa = 10.68AIGVLATSALILAGCGSTGGEE25 pKa = 3.79EE26 pKa = 4.15SEE28 pKa = 4.9PIVIGGKK35 pKa = 8.9PWTEE39 pKa = 4.0QYY41 pKa = 10.81ILPHH45 pKa = 7.07ILGQYY50 pKa = 9.7IEE52 pKa = 5.03ANSDD56 pKa = 3.49YY57 pKa = 10.43TVEE60 pKa = 4.22YY61 pKa = 10.34EE62 pKa = 4.31DD63 pKa = 4.27GLGEE67 pKa = 4.39VSILTPALEE76 pKa = 4.14QGDD79 pKa = 3.22IDD81 pKa = 6.02LYY83 pKa = 11.36VEE85 pKa = 4.11YY86 pKa = 9.92TGTGLKK92 pKa = 10.37DD93 pKa = 3.33VLKK96 pKa = 10.72QEE98 pKa = 4.44SEE100 pKa = 4.16PGQSSEE106 pKa = 4.38EE107 pKa = 3.72VLQRR111 pKa = 11.84VRR113 pKa = 11.84EE114 pKa = 4.56GYY116 pKa = 10.24EE117 pKa = 3.78EE118 pKa = 4.14EE119 pKa = 5.86LNATWLEE126 pKa = 3.82PLGFEE131 pKa = 4.79NGYY134 pKa = 7.77TLAYY138 pKa = 10.14SKK140 pKa = 11.15DD141 pKa = 3.36SGYY144 pKa = 10.96DD145 pKa = 3.08AEE147 pKa = 4.91TYY149 pKa = 11.0SDD151 pKa = 3.91LAEE154 pKa = 4.29ISQTEE159 pKa = 4.27DD160 pKa = 2.94MSFGAPHH167 pKa = 7.25PFYY170 pKa = 10.73EE171 pKa = 4.37RR172 pKa = 11.84QGDD175 pKa = 4.35GYY177 pKa = 11.06DD178 pKa = 4.39DD179 pKa = 6.31LIATYY184 pKa = 9.54PFEE187 pKa = 5.16FSATDD192 pKa = 3.43SFDD195 pKa = 3.54PAIMYY200 pKa = 7.64EE201 pKa = 4.0AVNNGEE207 pKa = 4.01VDD209 pKa = 4.12VIPAFTTDD217 pKa = 2.92SRR219 pKa = 11.84IGLFDD224 pKa = 5.22LEE226 pKa = 4.5TTEE229 pKa = 5.62DD230 pKa = 3.96DD231 pKa = 4.31LSFFPKK237 pKa = 9.98YY238 pKa = 10.1DD239 pKa = 3.42AAPVVRR245 pKa = 11.84LEE247 pKa = 4.41ALEE250 pKa = 4.09QYY252 pKa = 10.55PEE254 pKa = 4.37LEE256 pKa = 4.11SLLNEE261 pKa = 4.02LAGQISEE268 pKa = 4.42EE269 pKa = 4.1EE270 pKa = 4.08MLAMNARR277 pKa = 11.84VDD279 pKa = 3.41IDD281 pKa = 3.25QDD283 pKa = 3.68QPQDD287 pKa = 3.03IARR290 pKa = 11.84DD291 pKa = 3.63FLIEE295 pKa = 4.06KK296 pKa = 10.46GLIEE300 pKa = 4.14EE301 pKa = 4.41
MM1 pKa = 7.42KK2 pKa = 10.13KK3 pKa = 10.15KK4 pKa = 10.68AIGVLATSALILAGCGSTGGEE25 pKa = 3.79EE26 pKa = 4.15SEE28 pKa = 4.9PIVIGGKK35 pKa = 8.9PWTEE39 pKa = 4.0QYY41 pKa = 10.81ILPHH45 pKa = 7.07ILGQYY50 pKa = 9.7IEE52 pKa = 5.03ANSDD56 pKa = 3.49YY57 pKa = 10.43TVEE60 pKa = 4.22YY61 pKa = 10.34EE62 pKa = 4.31DD63 pKa = 4.27GLGEE67 pKa = 4.39VSILTPALEE76 pKa = 4.14QGDD79 pKa = 3.22IDD81 pKa = 6.02LYY83 pKa = 11.36VEE85 pKa = 4.11YY86 pKa = 9.92TGTGLKK92 pKa = 10.37DD93 pKa = 3.33VLKK96 pKa = 10.72QEE98 pKa = 4.44SEE100 pKa = 4.16PGQSSEE106 pKa = 4.38EE107 pKa = 3.72VLQRR111 pKa = 11.84VRR113 pKa = 11.84EE114 pKa = 4.56GYY116 pKa = 10.24EE117 pKa = 3.78EE118 pKa = 4.14EE119 pKa = 5.86LNATWLEE126 pKa = 3.82PLGFEE131 pKa = 4.79NGYY134 pKa = 7.77TLAYY138 pKa = 10.14SKK140 pKa = 11.15DD141 pKa = 3.36SGYY144 pKa = 10.96DD145 pKa = 3.08AEE147 pKa = 4.91TYY149 pKa = 11.0SDD151 pKa = 3.91LAEE154 pKa = 4.29ISQTEE159 pKa = 4.27DD160 pKa = 2.94MSFGAPHH167 pKa = 7.25PFYY170 pKa = 10.73EE171 pKa = 4.37RR172 pKa = 11.84QGDD175 pKa = 4.35GYY177 pKa = 11.06DD178 pKa = 4.39DD179 pKa = 6.31LIATYY184 pKa = 9.54PFEE187 pKa = 5.16FSATDD192 pKa = 3.43SFDD195 pKa = 3.54PAIMYY200 pKa = 7.64EE201 pKa = 4.0AVNNGEE207 pKa = 4.01VDD209 pKa = 4.12VIPAFTTDD217 pKa = 2.92SRR219 pKa = 11.84IGLFDD224 pKa = 5.22LEE226 pKa = 4.5TTEE229 pKa = 5.62DD230 pKa = 3.96DD231 pKa = 4.31LSFFPKK237 pKa = 9.98YY238 pKa = 10.1DD239 pKa = 3.42AAPVVRR245 pKa = 11.84LEE247 pKa = 4.41ALEE250 pKa = 4.09QYY252 pKa = 10.55PEE254 pKa = 4.37LEE256 pKa = 4.11SLLNEE261 pKa = 4.02LAGQISEE268 pKa = 4.42EE269 pKa = 4.1EE270 pKa = 4.08MLAMNARR277 pKa = 11.84VDD279 pKa = 3.41IDD281 pKa = 3.25QDD283 pKa = 3.68QPQDD287 pKa = 3.03IARR290 pKa = 11.84DD291 pKa = 3.63FLIEE295 pKa = 4.06KK296 pKa = 10.46GLIEE300 pKa = 4.14EE301 pKa = 4.41
Molecular weight: 33.39 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A365L8K6|A0A365L8K6_9BACL DNA mismatch repair protein MutT OS=Planococcus halotolerans OX=2233542 GN=DP120_03900 PE=4 SV=1
MM1 pKa = 7.61TLRR4 pKa = 11.84TYY6 pKa = 10.57QPNKK10 pKa = 8.74RR11 pKa = 11.84KK12 pKa = 9.54HH13 pKa = 5.99SKK15 pKa = 8.91VHH17 pKa = 5.68GFRR20 pKa = 11.84ARR22 pKa = 11.84MSTKK26 pKa = 9.71NGRR29 pKa = 11.84RR30 pKa = 11.84VIAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 8.8GRR40 pKa = 11.84KK41 pKa = 8.75VLSAA45 pKa = 4.05
MM1 pKa = 7.61TLRR4 pKa = 11.84TYY6 pKa = 10.57QPNKK10 pKa = 8.74RR11 pKa = 11.84KK12 pKa = 9.54HH13 pKa = 5.99SKK15 pKa = 8.91VHH17 pKa = 5.68GFRR20 pKa = 11.84ARR22 pKa = 11.84MSTKK26 pKa = 9.71NGRR29 pKa = 11.84RR30 pKa = 11.84VIAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 8.8GRR40 pKa = 11.84KK41 pKa = 8.75VLSAA45 pKa = 4.05
Molecular weight: 5.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1025973 |
34 |
2657 |
289.3 |
32.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.008 ± 0.043 | 0.56 ± 0.011 |
5.395 ± 0.034 | 8.087 ± 0.055 |
4.602 ± 0.035 | 7.139 ± 0.039 |
1.971 ± 0.02 | 7.305 ± 0.038 |
6.151 ± 0.034 | 9.679 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.936 ± 0.021 | 4.008 ± 0.029 |
3.685 ± 0.023 | 3.55 ± 0.025 |
4.204 ± 0.033 | 5.986 ± 0.029 |
5.386 ± 0.036 | 6.922 ± 0.03 |
1.05 ± 0.014 | 3.374 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
